当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
RLpMIEC: High-Affinity Peptide Generation Targeting Major Histocompatibility Complex-I Guided and Interpreted by Interaction Spectrum-Navigated Reinforcement Learning
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-08 , DOI: 10.1021/acs.jcim.4c01153 Qirui Deng 1 , Zhe Wang 2, 3 , Sutong Xiang 1 , Qinghua Wang 1 , Yifei Liu 2 , Tingjun Hou 2 , Huiyong Sun 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-08 , DOI: 10.1021/acs.jcim.4c01153 Qirui Deng 1 , Zhe Wang 2, 3 , Sutong Xiang 1 , Qinghua Wang 1 , Yifei Liu 2 , Tingjun Hou 2 , Huiyong Sun 1
Affiliation
|
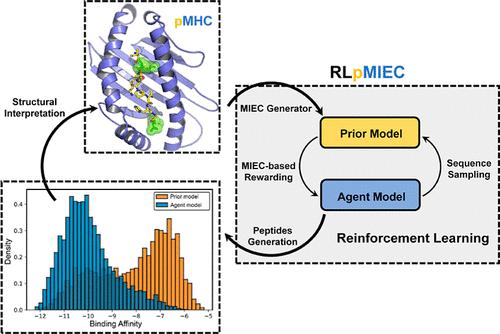
Major histocompatibility complex (MHC) plays a vital role in presenting epitopes (short peptides from pathogenic proteins) to T-cell receptors (TCRs) to trigger the subsequent immune responses. Vaccine design targeting MHC generally aims to find epitopes with a high binding affinity for MHC presentation. Nevertheless, to find novel epitopes usually requires high-throughput screening of bulk peptide database, which is time-consuming, labor-intensive, more unaffordable, and very expensive. Excitingly, the past several years have witnessed the great success of artificial intelligence (AI) in various fields, such as natural language processing (NLP, e.g., GPT-4), protein structure prediction and engineering (e.g., AlphaFold2), and so on. Therefore, herein, we propose a deep reinforcement-learning (RL)-based generative algorithm, RLpMIEC, to quantitatively design peptide targeting MHC-I systems. Specifically, RLpMIEC combines the energetic spectrum (namely, the molecular interaction energy component, MIEC) based on the peptide–MHC interaction and the sequence information to generate peptides with strong binding affinity and precise MIEC spectra to accelerate the discovery of candidate peptide vaccines. RLpMIEC performs well in all the generative capability evaluations and can generate peptides with strong binding affinities and precise MIECs and, moreover, with high interpretability, demonstrating its powerful capability in participation for accelerating peptide-based vaccine development.
中文翻译:

RLpMIEC:靶向主要组织相容性复合物 I 的高亲和力肽生成,由交互谱导航强化学习指导和解释
主要组织相容性复合体 (MHC) 在将表位(来自致病蛋白的短肽)呈递给 T 细胞受体 (TCR) 以触发后续免疫反应方面起着至关重要的作用。靶向 MHC 的疫苗设计通常旨在寻找对 MHC 呈递具有高结合亲和力的表位。然而,要找到新的表位,通常需要对大量多肽数据库进行高通量筛选,这很耗时、劳动密集、更负担不起且非常昂贵。令人兴奋的是,过去几年见证了人工智能 (AI) 在各个领域的巨大成功,例如自然语言处理(NLP,例如 GPT-4)、蛋白质结构预测和工程(例如 AlphaFold2)等。因此,在此,我们提出了一种基于深度强化学习 (RL) 的生成算法 RLpMIEC,用于定量设计靶向肽的 MHC-I 系统。具体来说,RLpMIEC 将基于肽-MHC 相互作用的能量谱(即分子相互作用能分量,MIEC)和序列信息相结合,生成具有强结合亲和力和精确 MIEC 光谱的肽,以加速候选肽疫苗的发现。RLpMIEC 在所有生成能力评估中都表现良好,可以生成具有强结合亲和力和精确 MIEC 的多肽,而且具有很高的可解释性,展示了其参与加速基于肽的疫苗开发的强大能力。
更新日期:2024-08-08
中文翻译:

RLpMIEC:靶向主要组织相容性复合物 I 的高亲和力肽生成,由交互谱导航强化学习指导和解释
主要组织相容性复合体 (MHC) 在将表位(来自致病蛋白的短肽)呈递给 T 细胞受体 (TCR) 以触发后续免疫反应方面起着至关重要的作用。靶向 MHC 的疫苗设计通常旨在寻找对 MHC 呈递具有高结合亲和力的表位。然而,要找到新的表位,通常需要对大量多肽数据库进行高通量筛选,这很耗时、劳动密集、更负担不起且非常昂贵。令人兴奋的是,过去几年见证了人工智能 (AI) 在各个领域的巨大成功,例如自然语言处理(NLP,例如 GPT-4)、蛋白质结构预测和工程(例如 AlphaFold2)等。因此,在此,我们提出了一种基于深度强化学习 (RL) 的生成算法 RLpMIEC,用于定量设计靶向肽的 MHC-I 系统。具体来说,RLpMIEC 将基于肽-MHC 相互作用的能量谱(即分子相互作用能分量,MIEC)和序列信息相结合,生成具有强结合亲和力和精确 MIEC 光谱的肽,以加速候选肽疫苗的发现。RLpMIEC 在所有生成能力评估中都表现良好,可以生成具有强结合亲和力和精确 MIEC 的多肽,而且具有很高的可解释性,展示了其参与加速基于肽的疫苗开发的强大能力。































 京公网安备 11010802027423号
京公网安备 11010802027423号