Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Comparative single-cell analyses identify shared and divergent features of human and mouse kidney development
Developmental Cell ( IF 10.7 ) Pub Date : 2024-08-08 , DOI: 10.1016/j.devcel.2024.07.013
Sunghyun Kim 1 , Kari Koppitch 1 , Riana K Parvez 1 , Jinjin Guo 1 , MaryAnne Achieng 1 , Jack Schnell 1 , Nils O Lindström 1 , Andrew P McMahon 1
Developmental Cell ( IF 10.7 ) Pub Date : 2024-08-08 , DOI: 10.1016/j.devcel.2024.07.013
Sunghyun Kim 1 , Kari Koppitch 1 , Riana K Parvez 1 , Jinjin Guo 1 , MaryAnne Achieng 1 , Jack Schnell 1 , Nils O Lindström 1 , Andrew P McMahon 1
Affiliation
|
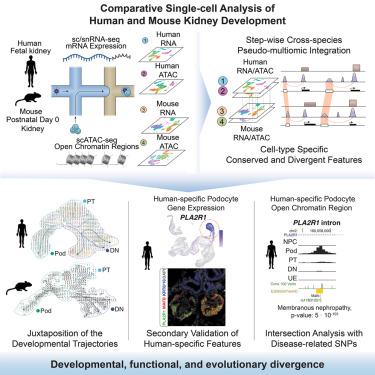
The mammalian kidney maintains fluid homeostasis through diverse epithelial cell types generated from nephron and ureteric progenitor cells. To extend a developmental understanding of the kidney’s epithelial networks, we compared chromatin organization (single-nuclear assay for transposase-accessible chromatin sequencing [ATAC-seq]; 112,864 nuclei) and gene expression (single-cell/nuclear RNA sequencing [RNA-seq]; 109,477 cells/nuclei) in the developing human (10.6–17.6 weeks; n = 10) and mouse (post-natal day [P]0; n = 10) kidney, supplementing analysis with published mouse datasets from earlier stages. Single-cell/nuclear datasets were analyzed at a species level, and then nephron and ureteric cellular lineages were extracted and integrated into a common, cross-species, multimodal dataset. Comparative computational analyses identified conserved and divergent features of chromatin organization and linked gene activity, identifying species-specific and cell-type-specific regulatory programs. In situ validation of human-enriched gene activity points to human-specific signaling interactions in kidney development. Further, human-specific enhancer regions were linked to kidney diseases through genome-wide association studies (GWASs), highlighting the potential for clinical insight from developmental modeling.
中文翻译:

比较单细胞分析确定了人和小鼠肾脏发育的共同特征和不同特征
哺乳动物肾脏通过肾单位和输尿管祖细胞产生的多种上皮细胞类型维持液体稳态。为了扩展对肾脏上皮网络的发育理解,我们比较了发育中的人类(10.6-17.6 周;n = 10)和小鼠(出生后 [P]0;n = 10)肾脏中的染色质组织(转座酶可及染色质测序的单核测定 [ATAC-seq];112,864 个细胞核)和基因表达(单细胞/核 RNA 测序 [RNA-seq];109,477 个细胞/核)肾脏,补充了早期已发表的小鼠数据集的分析。在物种水平上分析单细胞/核数据集,然后提取肾单位和输尿管细胞谱系并将其整合到一个通用的、跨物种的、多模式数据集中。比较计算分析确定了染色质组织和相关基因活性的保守和不同特征,确定了物种特异性和细胞类型特异性调节程序。人类富集基因活性的原位验证表明肾脏发育中的人类特异性信号相互作用。此外,人类特异性增强子区域通过全基因组关联研究 (GWAS) 与肾脏疾病相关联,突出了从发育建模中获得临床见解的潜力。
更新日期:2024-08-08
中文翻译:

比较单细胞分析确定了人和小鼠肾脏发育的共同特征和不同特征
哺乳动物肾脏通过肾单位和输尿管祖细胞产生的多种上皮细胞类型维持液体稳态。为了扩展对肾脏上皮网络的发育理解,我们比较了发育中的人类(10.6-17.6 周;n = 10)和小鼠(出生后 [P]0;n = 10)肾脏中的染色质组织(转座酶可及染色质测序的单核测定 [ATAC-seq];112,864 个细胞核)和基因表达(单细胞/核 RNA 测序 [RNA-seq];109,477 个细胞/核)肾脏,补充了早期已发表的小鼠数据集的分析。在物种水平上分析单细胞/核数据集,然后提取肾单位和输尿管细胞谱系并将其整合到一个通用的、跨物种的、多模式数据集中。比较计算分析确定了染色质组织和相关基因活性的保守和不同特征,确定了物种特异性和细胞类型特异性调节程序。人类富集基因活性的原位验证表明肾脏发育中的人类特异性信号相互作用。此外,人类特异性增强子区域通过全基因组关联研究 (GWAS) 与肾脏疾病相关联,突出了从发育建模中获得临床见解的潜力。

































 京公网安备 11010802027423号
京公网安备 11010802027423号