当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mask-Guided Target Node Feature Learning and Dynamic Detailed Feature Enhancement for lncRNA-Disease Association Prediction
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-07 , DOI: 10.1021/acs.jcim.4c00652 Ping Xuan 1, 2 , Wei Wang 1 , Hui Cui 3 , Shuai Wang 4 , Toshiya Nakaguchi 5 , Tiangang Zhang 2
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-07 , DOI: 10.1021/acs.jcim.4c00652 Ping Xuan 1, 2 , Wei Wang 1 , Hui Cui 3 , Shuai Wang 4 , Toshiya Nakaguchi 5 , Tiangang Zhang 2
Affiliation
|
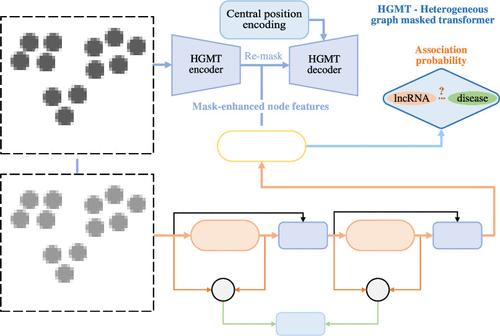
Identifying new relevant long noncoding RNAs (lncRNAs) for various human diseases can facilitate the exploration of the causes and progression of these diseases. Recently, several graph inference methods have been proposed to predict disease-related lncRNAs by exploiting the topological structure and node attributes within graphs. However, these methods did not prioritize the target lncRNA and disease nodes over auxiliary nodes like miRNA nodes, potentially limiting their ability to fully utilize the features of the target nodes. We propose a new method, mask-guided target node feature learning and dynamic detailed feature enhancement for lncRNA-disease association prediction (MDLD), to enhance node feature learning for improved lncRNA-disease association prediction. First, we designed a heterogeneous graph masked transformer autoencoder to guide feature learning, focusing more on the features of target lncRNA (disease) nodes. The target nodes were increasingly masked as training progressed, which helps develop a more robust prediction model. Second, we developed a graph convolutional network with dynamic residuals (GCNDR) to learn and integrate the heterogeneous topology and features of all lncRNA, disease, and miRNA nodes. GCNDR employs an interlayer residual strategy and a residual evolution strategy to mitigate oversmoothing caused by multilayer graph convolution. The interlayer residual strategy estimates the importance of node features learned in the previous GCN encoding layer for nodes in the current encoding layer. Additionally, since there are dependencies in the importance of features of individual lncRNA (disease, miRNA) nodes across multiple encoding layers, a gated recurrent unit-based strategy is proposed to encode these dependencies. Finally, we designed a perspective-level attention mechanism to obtain more informative features of lncRNA and disease node pairs from the perspectives of mask-enhanced and dynamic-enhanced node features. Cross-validation experimental results demonstrated that MDLD outperformed 10 other state-of-the-art prediction methods. Ablation experiments and case studies on candidate lncRNAs for three diseases further proved the technical contributions of MDLD and its capability to discover disease-related lncRNAs.
中文翻译:

用于 lncRNA 疾病关联预测的掩模引导目标节点特征学习和动态详细特征增强
鉴定与各种人类疾病相关的新的长非编码RNA(lncRNA)可以促进探索这些疾病的原因和进展。最近,人们提出了几种图推理方法,通过利用图中的拓扑结构和节点属性来预测与疾病相关的lncRNA。然而,这些方法并没有将目标 lncRNA 和疾病节点优先于 miRNA 节点等辅助节点,这可能限制了它们充分利用目标节点特征的能力。我们提出了一种新方法,即掩模引导的目标节点特征学习和lncRNA-疾病关联预测(MDLD)的动态详细特征增强,以增强节点特征学习以改进lncRNA-疾病关联预测。首先,我们设计了一个异构图掩码变压器自动编码器来指导特征学习,更多地关注目标lncRNA(疾病)节点的特征。随着训练的进行,目标节点越来越被掩盖,这有助于开发更稳健的预测模型。其次,我们开发了一个具有动态残差的图卷积网络(GCNDR)来学习和集成所有 lncRNA、疾病和 miRNA 节点的异构拓扑和特征。 GCNDR采用层间残差策略和残差进化策略来减轻多层图卷积引起的过度平滑。层间残差策略估计前一个GCN编码层中学习到的节点特征对于当前编码层中的节点的重要性。此外,由于跨多个编码层的各个lncRNA(疾病,miRNA)节点的特征的重要性存在依赖性,因此提出了基于门控循环单元的策略来编码这些依赖性。 最后,我们设计了一种透视级注意力机制,从掩模增强和动态增强节点特征的角度获得lncRNA和疾病节点对的更多信息特征。交叉验证实验结果表明,MDLD 优于其他 10 种最先进的预测方法。针对三种疾病候选lncRNA的消融实验和案例研究进一步证明了MDLD的技术贡献及其发现疾病相关lncRNA的能力。
更新日期:2024-08-07
中文翻译:

用于 lncRNA 疾病关联预测的掩模引导目标节点特征学习和动态详细特征增强
鉴定与各种人类疾病相关的新的长非编码RNA(lncRNA)可以促进探索这些疾病的原因和进展。最近,人们提出了几种图推理方法,通过利用图中的拓扑结构和节点属性来预测与疾病相关的lncRNA。然而,这些方法并没有将目标 lncRNA 和疾病节点优先于 miRNA 节点等辅助节点,这可能限制了它们充分利用目标节点特征的能力。我们提出了一种新方法,即掩模引导的目标节点特征学习和lncRNA-疾病关联预测(MDLD)的动态详细特征增强,以增强节点特征学习以改进lncRNA-疾病关联预测。首先,我们设计了一个异构图掩码变压器自动编码器来指导特征学习,更多地关注目标lncRNA(疾病)节点的特征。随着训练的进行,目标节点越来越被掩盖,这有助于开发更稳健的预测模型。其次,我们开发了一个具有动态残差的图卷积网络(GCNDR)来学习和集成所有 lncRNA、疾病和 miRNA 节点的异构拓扑和特征。 GCNDR采用层间残差策略和残差进化策略来减轻多层图卷积引起的过度平滑。层间残差策略估计前一个GCN编码层中学习到的节点特征对于当前编码层中的节点的重要性。此外,由于跨多个编码层的各个lncRNA(疾病,miRNA)节点的特征的重要性存在依赖性,因此提出了基于门控循环单元的策略来编码这些依赖性。 最后,我们设计了一种透视级注意力机制,从掩模增强和动态增强节点特征的角度获得lncRNA和疾病节点对的更多信息特征。交叉验证实验结果表明,MDLD 优于其他 10 种最先进的预测方法。针对三种疾病候选lncRNA的消融实验和案例研究进一步证明了MDLD的技术贡献及其发现疾病相关lncRNA的能力。































 京公网安备 11010802027423号
京公网安备 11010802027423号