当前位置:
X-MOL 学术
›
Mol. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
In vivo assembly of bacterial partition condensates on circular supercoiled and linear DNA
Molecular Microbiology ( IF 2.6 ) Pub Date : 2024-08-07 , DOI: 10.1111/mmi.15297 Hicham Sekkouri Alaoui 1 , Valentin Quèbre 1 , Linda Delimi 2 , Jérôme Rech 1 , Roxanne Debaugny-Diaz 1 , Delphine Labourdette 3 , Manuel Campos 1 , François Cornet 1 , Jean-Charles Walter 2 , Jean-Yves Bouet 1
Molecular Microbiology ( IF 2.6 ) Pub Date : 2024-08-07 , DOI: 10.1111/mmi.15297 Hicham Sekkouri Alaoui 1 , Valentin Quèbre 1 , Linda Delimi 2 , Jérôme Rech 1 , Roxanne Debaugny-Diaz 1 , Delphine Labourdette 3 , Manuel Campos 1 , François Cornet 1 , Jean-Charles Walter 2 , Jean-Yves Bouet 1
Affiliation
|
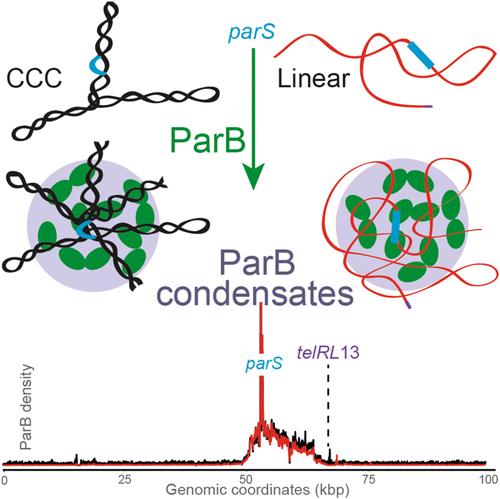
In bacteria, faithful DNA segregation of chromosomes and plasmids is mainly mediated by ParABS systems. These systems, consisting of a ParA ATPase, a DNA binding ParB CTPase, and centromere sites parS , orchestrate the separation of newly replicated DNA copies and their intracellular positioning. Accurate segregation relies on the assembly of a high‐molecular‐weight complex, comprising a few hundreds of ParB dimers nucleated from parS sites. This complex assembles in a multi‐step process and exhibits dynamic liquid‐droplet properties. Despite various proposed models, the complete mechanism for partition complex assembly remains elusive. This study investigates the impact of DNA supercoiling on ParB DNA binding profiles in vivo, using the ParABS system of the plasmid F. We found that variations in DNA supercoiling does not significantly affect any steps in the assembly of the partition complex. Furthermore, physical modeling, leveraging ChIP‐seq data from linear plasmids F, suggests that ParB sliding is restricted to approximately 2 Kbp from parS , highlighting the necessity for additional mechanisms beyond ParB sliding over DNA for concentrating ParB into condensates nucleated at parS . Finally, explicit simulations of a polymer coated with bound ParB suggest a dominant role for ParB‐ParB interactions in DNA compaction within ParB condensates.
中文翻译:

圆形超螺旋和线性 DNA 上细菌分区凝聚物的体内组装
在细菌中,染色体和质粒的 DNA 忠实分离主要由 ParABS 系统介导。这些系统由 ParA ATP 酶、DNA 结合 ParB CTP 酶和着丝粒位点组成帕斯,协调新复制的 DNA 拷贝的分离及其细胞内定位。准确的分离依赖于高分子量复合物的组装,该复合物由数百个 ParB 二聚体组成帕斯网站。这种复合物通过多步骤过程组装并表现出动态液滴特性。尽管提出了各种模型,但分区复杂组装的完整机制仍然难以捉摸。本研究使用质粒 F 的 ParABS 系统研究了 DNA 超螺旋对 ParB DNA 体内结合谱的影响。我们发现 DNA 超螺旋的变化不会显着影响分区复合物组装的任何步骤。此外,利用线性质粒 F 的 ChIP-seq 数据进行的物理建模表明,ParB 滑动限制在大约 2 Kbp帕斯,强调了除了 ParB 在 DNA 上滑动之外还需要其他机制的必要性,以便将 ParB 浓缩成在帕斯。最后,对涂有结合 ParB 的聚合物的显式模拟表明,ParB-ParB 相互作用在 ParB 凝聚物内的 DNA 压缩中起主导作用。
更新日期:2024-08-07
中文翻译:

圆形超螺旋和线性 DNA 上细菌分区凝聚物的体内组装
在细菌中,染色体和质粒的 DNA 忠实分离主要由 ParABS 系统介导。这些系统由 ParA ATP 酶、DNA 结合 ParB CTP 酶和着丝粒位点组成帕斯,协调新复制的 DNA 拷贝的分离及其细胞内定位。准确的分离依赖于高分子量复合物的组装,该复合物由数百个 ParB 二聚体组成帕斯网站。这种复合物通过多步骤过程组装并表现出动态液滴特性。尽管提出了各种模型,但分区复杂组装的完整机制仍然难以捉摸。本研究使用质粒 F 的 ParABS 系统研究了 DNA 超螺旋对 ParB DNA 体内结合谱的影响。我们发现 DNA 超螺旋的变化不会显着影响分区复合物组装的任何步骤。此外,利用线性质粒 F 的 ChIP-seq 数据进行的物理建模表明,ParB 滑动限制在大约 2 Kbp帕斯,强调了除了 ParB 在 DNA 上滑动之外还需要其他机制的必要性,以便将 ParB 浓缩成在帕斯。最后,对涂有结合 ParB 的聚合物的显式模拟表明,ParB-ParB 相互作用在 ParB 凝聚物内的 DNA 压缩中起主导作用。






























 京公网安备 11010802027423号
京公网安备 11010802027423号