当前位置:
X-MOL 学术
›
Environ. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Microdiversity in marine pelagic ammonia‐oxidizing archaeal populations in a Mediterranean long‐read metagenome
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-07-31 , DOI: 10.1111/1462-2920.16684 Pablo Suárez-Moo 1 , Jose M Haro-Moreno 1 , Francisco Rodriguez-Valera 1
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-07-31 , DOI: 10.1111/1462-2920.16684 Pablo Suárez-Moo 1 , Jose M Haro-Moreno 1 , Francisco Rodriguez-Valera 1
Affiliation
|
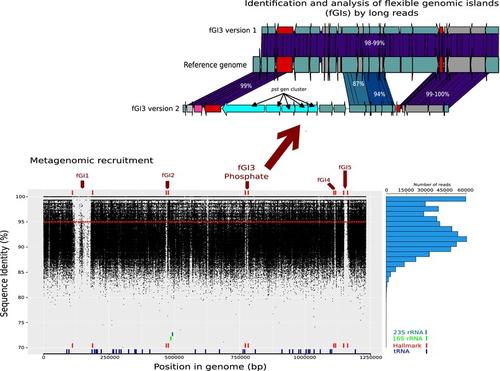
The knowledge of the different population‐level processes operating within a species, and the genetic variability of the individual prokaryotic genomes, is key to understanding the adaptability of microbial populations. Here, we characterized the flexible genome of ammonia‐oxidizing archaeal (AOA) populations using a metagenomic recruitment approach and long‐read (PacBio HiFi) metagenomic sequencing. In the lower photic zone of the western Mediterranean Sea (75 m deep), the genomes Nitrosopelagicus brevis CN25 and Nitrosopumilus catalinensis SPOT1 had the highest recruitment values among available complete AOA genomes. They were used to analyse the diversity of flexible genes (variable from strain to strain) by examining the long‐reads located within the flexible genomic islands (fGIs) identified by their under‐recruitment. Both AOA genomes had a large fGI involved in the glycosylation of exposed structures, highly variable, and rich in glycosyltransferases. N. brevis had two fGIs related to the transport of phosphorus and ammonium respectively. N. catalinensis had fGIs involved in phosphorus transportation and metal uptake. A fGI5 previously reported as ‘unassigned function’ in N. brevis could be associated with defense. These findings demonstrate that the microdiversity of marine microbe populations, including AOA, can be effectively characterized using an approach that incorporates third‐generation sequencing metagenomics.
中文翻译:

地中海长读元基因组中海洋中上层氨氧化古菌种群的微多样性
了解一个物种内运行的不同种群水平过程以及单个原核基因组的遗传变异性是了解微生物种群适应性的关键。在这里,我们使用宏基因组招募方法和长读长(PacBio HiFi)宏基因组测序来表征氨氧化古菌(AOA)群体的灵活基因组。在地中海西部的低透光区(75 m深),基因组短亚硝基鱼CN25 和加泰罗尼亚亚硝草SPOT1 在可用的完整 AOA 基因组中具有最高的招募值。它们被用来通过检查位于灵活基因组岛(fGI)内的长读段来分析灵活基因的多样性(因菌株而异)。两个 AOA 基因组都有一个很大的 fGI,参与暴露结构的糖基化,高度可变,并且富含糖基转移酶。短猪笼草有两个 fGI 分别与磷和铵的运输有关。加泰罗尼亚猪笼草fGI 参与磷运输和金属吸收。 fGI5 之前被报告为“未分配的功能”短猪笼草可能与防御有关。这些发现表明,使用结合第三代测序宏基因组学的方法可以有效地表征包括 AOA 在内的海洋微生物种群的微观多样性。
更新日期:2024-07-31
中文翻译:

地中海长读元基因组中海洋中上层氨氧化古菌种群的微多样性
了解一个物种内运行的不同种群水平过程以及单个原核基因组的遗传变异性是了解微生物种群适应性的关键。在这里,我们使用宏基因组招募方法和长读长(PacBio HiFi)宏基因组测序来表征氨氧化古菌(AOA)群体的灵活基因组。在地中海西部的低透光区(75 m深),基因组短亚硝基鱼CN25 和加泰罗尼亚亚硝草SPOT1 在可用的完整 AOA 基因组中具有最高的招募值。它们被用来通过检查位于灵活基因组岛(fGI)内的长读段来分析灵活基因的多样性(因菌株而异)。两个 AOA 基因组都有一个很大的 fGI,参与暴露结构的糖基化,高度可变,并且富含糖基转移酶。短猪笼草有两个 fGI 分别与磷和铵的运输有关。加泰罗尼亚猪笼草fGI 参与磷运输和金属吸收。 fGI5 之前被报告为“未分配的功能”短猪笼草可能与防御有关。这些发现表明,使用结合第三代测序宏基因组学的方法可以有效地表征包括 AOA 在内的海洋微生物种群的微观多样性。































 京公网安备 11010802027423号
京公网安备 11010802027423号