当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Transcriptome-Wide, Unbiased Profiling of Ribonuclease Targeting Chimeras
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2024-07-24 , DOI: 10.1021/jacs.4c04717 Yuquan Tong 1, 2 , Xiaoxuan Su 1, 2 , Warren Rouse 3 , Jessica L Childs-Disney 2 , Amirhossein Taghavi 2 , Patrick R A Zanon 2 , Sandra Kovachka 2 , Tenghui Wang 1, 2 , Walter N Moss 3 , Matthew D Disney 1, 2
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2024-07-24 , DOI: 10.1021/jacs.4c04717 Yuquan Tong 1, 2 , Xiaoxuan Su 1, 2 , Warren Rouse 3 , Jessica L Childs-Disney 2 , Amirhossein Taghavi 2 , Patrick R A Zanon 2 , Sandra Kovachka 2 , Tenghui Wang 1, 2 , Walter N Moss 3 , Matthew D Disney 1, 2
Affiliation
|
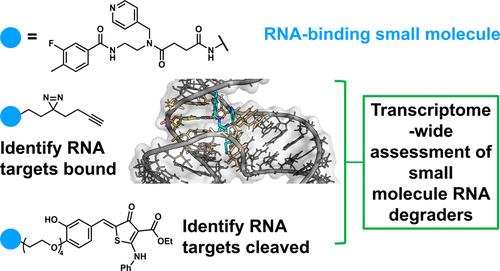
Various approaches have been developed to target RNA and modulate its function with modes of action including binding and cleavage. Herein, we explored how small molecule binding is correlated with cleavage induced by heterobifunctional ribonuclease targeting chimeras (RiboTACs), where RNase L is recruited to cleave the bound RNA target, in a transcriptome-wide, unbiased fashion. Only a fraction of bound targets was cleaved by RNase L, induced by RiboTAC binding. Global analysis suggested that (i) cleaved targets generally form a region of stable structure that encompasses the small molecule binding site; (ii) cleaved targets have preferred RNase L cleavage sites nearby small molecule binding sites; (iii) RiboTACs facilitate a cellular interaction between cleaved targets and RNase L; and (iv) the expression level of the target influences the extent of cleavage observed. In one example, we converted a binder of LGALS1 (galectin-1) mRNA into a RiboTAC. In MDA-MB-231 cells, the binder had no effect on galectin-1 protein levels, while the RiboTAC cleaved LGALS1 mRNA, reduced galectin-1 protein abundance, and affected galectin-1-associated oncogenic cellular phenotypes. Using LGALS1, we further assessed additional factors including the length of the linker that tethers the two components of the RiboTAC, cellular uptake, and the RNase L-recruiting module on RiboTAC potency. Collectively, these studies may facilitate triangulation of factors to enable the design of RiboTACs.
中文翻译:

靶向嵌合体的核糖核酸酶的全转录组、无偏差分析
人们已经开发出多种方法来靶向 RNA 并通过结合和切割等作用模式调节其功能。在此,我们探讨了小分子结合如何与异双功能核糖核酸酶靶向嵌合体 (RiboTAC) 诱导的切割相关,其中 RNase L 被招募来以全转录组范围、无偏倚的方式切割结合的 RNA 靶标。只有一小部分结合的靶标被 RiboTAC 结合诱导的 RNase L 裂解。整体分析表明(i)切割的靶标通常形成包含小分子结合位点的稳定结构区域; (ii) 切割的靶标在小分子结合位点附近具有首选的 RNase L 切割位点; (iii) RiboTAC 促进切割靶标和 RNase L 之间的细胞相互作用; (iv) 靶标的表达水平影响观察到的切割程度。在一个示例中,我们将LGALS1 (galectin-1) mRNA 的结合物转化为 RiboTAC。在 MDA-MB-231 细胞中,结合剂对 galectin-1 蛋白水平没有影响,而 RiboTAC 切割LGALS1 mRNA,降低 galectin-1 蛋白丰度,并影响 galectin-1 相关的致癌细胞表型。使用LGALS1 ,我们进一步评估了其他因素,包括连接 RiboTAC 两个组件的接头长度、细胞摄取以及 RiboTAC 效力上的 RNase L 募集模块。总的来说,这些研究可能有助于对因素进行三角测量,以实现 RiboTAC 的设计。
更新日期:2024-07-24
中文翻译:

靶向嵌合体的核糖核酸酶的全转录组、无偏差分析
人们已经开发出多种方法来靶向 RNA 并通过结合和切割等作用模式调节其功能。在此,我们探讨了小分子结合如何与异双功能核糖核酸酶靶向嵌合体 (RiboTAC) 诱导的切割相关,其中 RNase L 被招募来以全转录组范围、无偏倚的方式切割结合的 RNA 靶标。只有一小部分结合的靶标被 RiboTAC 结合诱导的 RNase L 裂解。整体分析表明(i)切割的靶标通常形成包含小分子结合位点的稳定结构区域; (ii) 切割的靶标在小分子结合位点附近具有首选的 RNase L 切割位点; (iii) RiboTAC 促进切割靶标和 RNase L 之间的细胞相互作用; (iv) 靶标的表达水平影响观察到的切割程度。在一个示例中,我们将LGALS1 (galectin-1) mRNA 的结合物转化为 RiboTAC。在 MDA-MB-231 细胞中,结合剂对 galectin-1 蛋白水平没有影响,而 RiboTAC 切割LGALS1 mRNA,降低 galectin-1 蛋白丰度,并影响 galectin-1 相关的致癌细胞表型。使用LGALS1 ,我们进一步评估了其他因素,包括连接 RiboTAC 两个组件的接头长度、细胞摄取以及 RiboTAC 效力上的 RNase L 募集模块。总的来说,这些研究可能有助于对因素进行三角测量,以实现 RiboTAC 的设计。

















































 京公网安备 11010802027423号
京公网安备 11010802027423号