当前位置:
X-MOL 学术
›
J. Agric. Food Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Discovery Phase Agrochemical Predictive Safety Assessment Using High Content In Vitro Data to Estimate an In Vivo Toxicity Point of Departure
Journal of Agricultural and Food Chemistry ( IF 5.7 ) Pub Date : 2024-07-21 , DOI: 10.1021/acs.jafc.4c03094 Enrica Bianchi 1 , Eduardo Costa 2 , Joshua Harrill 3 , Paul Deford 1 , Jessica LaRocca 1 , Wei Chen 1 , Zachary Sutake 1 , Audrey Lehman 1 , Anthony Pappas-Garton 4 , Eric Sherer 1 , Connor Moreillon 1 , Shreedharan Sriram 1 , Andi Dhroso 1 , Kamin Johnson 1
Journal of Agricultural and Food Chemistry ( IF 5.7 ) Pub Date : 2024-07-21 , DOI: 10.1021/acs.jafc.4c03094 Enrica Bianchi 1 , Eduardo Costa 2 , Joshua Harrill 3 , Paul Deford 1 , Jessica LaRocca 1 , Wei Chen 1 , Zachary Sutake 1 , Audrey Lehman 1 , Anthony Pappas-Garton 4 , Eric Sherer 1 , Connor Moreillon 1 , Shreedharan Sriram 1 , Andi Dhroso 1 , Kamin Johnson 1
Affiliation
|
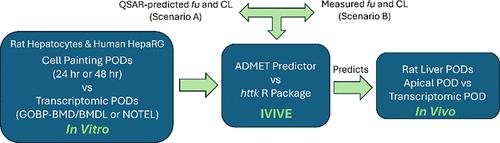
Utilization of in vitro (cellular) techniques, like Cell Painting and transcriptomics, could provide powerful tools for agrochemical candidate sorting and selection in the discovery process. However, using these models generates challenges translating in vitro concentrations to the corresponding in vivo exposures. Physiologically based pharmacokinetic (PBPK) modeling provides a framework for quantitative in vitro to in vivo extrapolation (IVIVE). We tested whether in vivo (rat liver) transcriptomic and apical points of departure (PODs) could be accurately predicted from in vitro (rat hepatocyte or human HepaRG) transcriptomic PODs or HepaRG Cell Painting PODs using PBPK modeling. We compared two PBPK models, the ADMET predictor and the httk R package, and found httk to predict the in vivo PODs more accurately. Our findings suggest that a rat liver apical and transcriptomic POD can be estimated utilizing a combination of in vitro transcriptome-based PODs coupled with PBPK modeling for IVIVE. Thus, high content in vitro data can be translated with modest accuracy to in vivo models of ultimate regulatory importance to help select agrochemical analogs in early stage discovery program.
中文翻译:

发现阶段农用化学品预测安全评估使用高含量体外数据来估计体内毒性出发点
利用体外(细胞)技术,如细胞涂色和转录组学,可以为发现过程中农用化学品候选物的分类和选择提供强大的工具。然而,使用这些模型会产生将体外浓度转化为相应的体内暴露量的挑战。基于生理学的药代动力学 (PBPK) 模型为定量体外到体内外推 (IVIVE) 提供了框架。我们测试了是否可以使用 PBPK 模型从体外(大鼠肝细胞或人 HepaRG)转录组 POD 或 HepaRG 细胞绘制 POD 准确预测体内(大鼠肝脏)转录组和顶端出发点 (POD)。我们比较了两个 PBPK 模型、ADMET 预测器和httk R 包,发现httk可以更准确地预测体内POD。我们的研究结果表明,可以利用基于体外转录组的 POD 与 IVIVE 的 PBPK 模型相结合来估计大鼠肝尖和转录组 POD。因此,高含量的体外数据可以以适度的精度转化为具有最终监管重要性的体内模型,以帮助在早期发现计划中选择农用化学品类似物。
更新日期:2024-07-21
中文翻译:

发现阶段农用化学品预测安全评估使用高含量体外数据来估计体内毒性出发点
利用体外(细胞)技术,如细胞涂色和转录组学,可以为发现过程中农用化学品候选物的分类和选择提供强大的工具。然而,使用这些模型会产生将体外浓度转化为相应的体内暴露量的挑战。基于生理学的药代动力学 (PBPK) 模型为定量体外到体内外推 (IVIVE) 提供了框架。我们测试了是否可以使用 PBPK 模型从体外(大鼠肝细胞或人 HepaRG)转录组 POD 或 HepaRG 细胞绘制 POD 准确预测体内(大鼠肝脏)转录组和顶端出发点 (POD)。我们比较了两个 PBPK 模型、ADMET 预测器和httk R 包,发现httk可以更准确地预测体内POD。我们的研究结果表明,可以利用基于体外转录组的 POD 与 IVIVE 的 PBPK 模型相结合来估计大鼠肝尖和转录组 POD。因此,高含量的体外数据可以以适度的精度转化为具有最终监管重要性的体内模型,以帮助在早期发现计划中选择农用化学品类似物。

















































 京公网安备 11010802027423号
京公网安备 11010802027423号