当前位置:
X-MOL 学术
›
Pest Manag. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Enhancing practicality of deep learning for crop disease identification under field conditions: insights from model evaluation and crop‐specific approaches
Pest Management Science ( IF 3.8 ) Pub Date : 2024-07-19 , DOI: 10.1002/ps.8317 Qi Tian 1 , Gang Zhao 2, 3 , Changqing Yan 4 , Linjia Yao 1 , Junjie Qu 5 , Ling Yin 5 , Hao Feng 2, 6 , Ning Yao 7 , Qiang Yu 2
Pest Management Science ( IF 3.8 ) Pub Date : 2024-07-19 , DOI: 10.1002/ps.8317 Qi Tian 1 , Gang Zhao 2, 3 , Changqing Yan 4 , Linjia Yao 1 , Junjie Qu 5 , Ling Yin 5 , Hao Feng 2, 6 , Ning Yao 7 , Qiang Yu 2
Affiliation
|
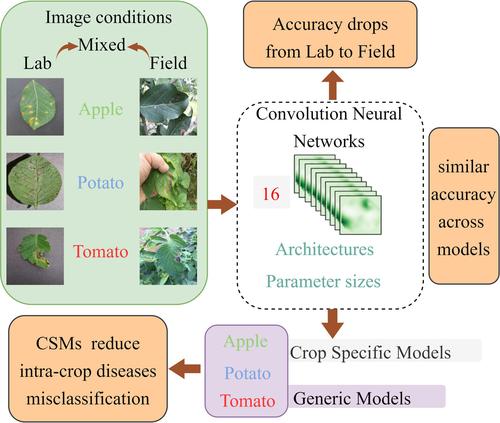
BACKGROUNDCrop diseases can lead to significant yield losses and food shortages if not promptly identified and managed by farmers. With the advancements in convolutional neural networks (CNN) and the widespread availability of smartphones, automated and accurate identification of crop diseases has become feasible. However, although previous studies have achieved high accuracy (>95%) under laboratory conditions (Lab) using mixed data sets of multiple crops, these models often falter when deployed under field conditions (Field). In this study, we aimed to evaluate disease identification accuracy under Lab, Field, and Mixed (Lab and Field) conditions using an assembled data set encompassing 14 diseases of apple (Malus × domestica Borkh.), potato (Solanum tuberosum L.), and tomato (Solanum lycopersicum L.). In addition, we investigated the impact of model architectures, parameter sizes, and crop‐specific models (CSMs) on accuracy, using DenseNets, ResNets, MobileNetV3, EfficientNet, and VGG Nets.RESULTSOur results revealed a decrease in accuracy across all models from Lab (98.22%) to Mixed (91.76%) to Field (71.55%) conditions. Interestingly, disease classification accuracy showed minimal variation across model architectures and parameter sizes: Lab (97.61–98.76%), Mixed (90.76–92.31%), and Field (68.56–73.81%). Although CSMs were found to reduce inter‐crop disease misclassifications, they also led to a slight increase in intra‐crop misclassifications.CONCLUSIONOur findings underscore the importance of enriching data representation and volumes over employing new model architectures. Furthermore, the need for more field‐specific images was highlighted. Ultimately, these insights contribute to the advancement of crop disease identification applications, facilitating their practical implementation in farmer's fields. © 2024 Society of Chemical Industry.
中文翻译:

增强田间条件下作物病害识别深度学习的实用性:来自模型评估和作物特定方法的见解
背景技术如果农民不及时识别和管理作物病害,可能会导致严重的产量损失和粮食短缺。随着卷积神经网络(CNN)的进步和智能手机的广泛使用,自动、准确地识别农作物病害已成为可能。然而,尽管之前的研究使用多种作物的混合数据集在实验室条件(Lab)下实现了高精度(>95%),但这些模型在田间条件(Field)下部署时往往会出现问题。在本研究中,我们旨在使用包含 14 种苹果病害的组装数据集,评估实验室、田间和混合(实验室和田间)条件下病害识别的准确性(海棠属×家畜Borkh。),土豆(马铃薯L.)和番茄(番茄L.)。此外,我们使用 DenseNets、ResNets、MobileNetV3、EfficientNet 和 VGG Nets 研究了模型架构、参数大小和作物特定模型 (CSM) 对准确性的影响。结果我们的结果显示,实验室所有模型的准确性均有所下降(98.22%) 到混合 (91.76%) 到现场 (71.55%) 条件。有趣的是,疾病分类准确性在模型架构和参数大小之间显示出最小的变化:实验室(97.61-98.76%)、混合(90.76-92.31%)和现场(68.56-73.81%)。尽管发现 CSM 可以减少作物间疾病错误分类,但它们也导致作物内错误分类略有增加。结论我们的研究结果强调了丰富数据表示和数据量比采用新模型架构的重要性。此外,还强调了对更多特定领域图像的需求。 最终,这些见解有助于推进作物病害识别应用,促进其在农民田间的实际实施。 © 2024 化学工业协会。
更新日期:2024-07-19
中文翻译:

增强田间条件下作物病害识别深度学习的实用性:来自模型评估和作物特定方法的见解
背景技术如果农民不及时识别和管理作物病害,可能会导致严重的产量损失和粮食短缺。随着卷积神经网络(CNN)的进步和智能手机的广泛使用,自动、准确地识别农作物病害已成为可能。然而,尽管之前的研究使用多种作物的混合数据集在实验室条件(Lab)下实现了高精度(>95%),但这些模型在田间条件(Field)下部署时往往会出现问题。在本研究中,我们旨在使用包含 14 种苹果病害的组装数据集,评估实验室、田间和混合(实验室和田间)条件下病害识别的准确性(海棠属×家畜Borkh。),土豆(马铃薯L.)和番茄(番茄L.)。此外,我们使用 DenseNets、ResNets、MobileNetV3、EfficientNet 和 VGG Nets 研究了模型架构、参数大小和作物特定模型 (CSM) 对准确性的影响。结果我们的结果显示,实验室所有模型的准确性均有所下降(98.22%) 到混合 (91.76%) 到现场 (71.55%) 条件。有趣的是,疾病分类准确性在模型架构和参数大小之间显示出最小的变化:实验室(97.61-98.76%)、混合(90.76-92.31%)和现场(68.56-73.81%)。尽管发现 CSM 可以减少作物间疾病错误分类,但它们也导致作物内错误分类略有增加。结论我们的研究结果强调了丰富数据表示和数据量比采用新模型架构的重要性。此外,还强调了对更多特定领域图像的需求。 最终,这些见解有助于推进作物病害识别应用,促进其在农民田间的实际实施。 © 2024 化学工业协会。











































 京公网安备 11010802027423号
京公网安备 11010802027423号