Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Crystal structure of lipase from Pseudomonas aeruginosa reveals an unusual catalytic triad conformation
Structure ( IF 4.4 ) Pub Date : 2024-07-17 , DOI: 10.1016/j.str.2024.06.014 Gang Xu 1 , Hua Guo 2 , Zhonglang Yu 1 , Shulin Wang 1 , Dandan Shen 1 , Lirong Yang 2 , Jianping Wu 2 , Binbin Chen 2 , Haoran Yu 2
Structure ( IF 4.4 ) Pub Date : 2024-07-17 , DOI: 10.1016/j.str.2024.06.014 Gang Xu 1 , Hua Guo 2 , Zhonglang Yu 1 , Shulin Wang 1 , Dandan Shen 1 , Lirong Yang 2 , Jianping Wu 2 , Binbin Chen 2 , Haoran Yu 2
Affiliation
|
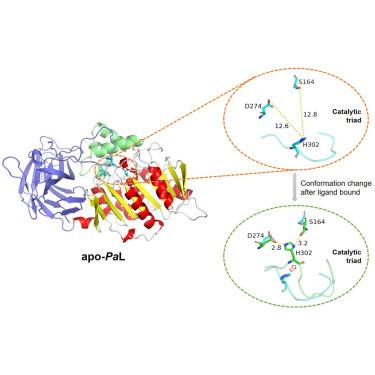
The Pseudomonas aeruginosa lipase Pa L catalyzes the stereoselective hydrolysis of menthyl propionate to produce L-menthol. The lack of a three-dimensional structure of Pa L has so far prevented a detailed understanding of its stereoselective reaction mechanism. Here, the crystal structure of Pa L was determined at a resolution of 1.80 Å by single-wavelength anomalous diffraction. In the apo-Pa L structure, the catalytic His302 is located in a long loop on the surface that is solvent exposed. His302 is distant from the other two catalytic residues, Asp274 and Ser164. This configuration of catalytic residues is unusual for lipases. Using metadynamics simulations, we observed that the enzyme undergoes a significant conformational change upon ligand binding. We also explored the catalytic and stereoselectivity mechanisms of Pa L by all-atom molecular dynamics simulations. These findings could guide the engineering of Pa L with an improved diastereoselectivity for L-menthol production.
中文翻译:

铜绿假单胞菌脂肪酶的晶体结构揭示了不寻常的催化三联体构象
铜绿假单胞菌脂肪酶PaL催化丙酸薄荷酯立体选择性水解产生L-薄荷醇。迄今为止,PaL 三维结构的缺乏阻碍了对其立体选择性反应机制的详细了解。在这里,通过单波长反常衍射以 1.80 Å 的分辨率测定了 PaL 的晶体结构。在 apo-PaL 结构中,催化 His3O2 位于暴露于溶剂的表面上的长环中。 His302 距离其他两个催化残基 Asp274 和 Ser164 较远。这种催化残基的构型对于脂肪酶来说是不常见的。使用元动力学模拟,我们观察到该酶在配体结合后经历了显着的构象变化。我们还通过全原子分子动力学模拟探索了 PaL 的催化和立体选择性机制。这些发现可以指导 PaL 的工程设计,提高 L-薄荷醇生产的非对映选择性。
更新日期:2024-07-17
中文翻译:

铜绿假单胞菌脂肪酶的晶体结构揭示了不寻常的催化三联体构象
铜绿假单胞菌脂肪酶PaL催化丙酸薄荷酯立体选择性水解产生L-薄荷醇。迄今为止,PaL 三维结构的缺乏阻碍了对其立体选择性反应机制的详细了解。在这里,通过单波长反常衍射以 1.80 Å 的分辨率测定了 PaL 的晶体结构。在 apo-PaL 结构中,催化 His3O2 位于暴露于溶剂的表面上的长环中。 His302 距离其他两个催化残基 Asp274 和 Ser164 较远。这种催化残基的构型对于脂肪酶来说是不常见的。使用元动力学模拟,我们观察到该酶在配体结合后经历了显着的构象变化。我们还通过全原子分子动力学模拟探索了 PaL 的催化和立体选择性机制。这些发现可以指导 PaL 的工程设计,提高 L-薄荷醇生产的非对映选择性。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号