当前位置:
X-MOL 学术
›
Environ. Sci. Technol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Simple Genomic Traits Predict Rates of Polysaccharide Biodegradation
Environmental Science & Technology ( IF 10.8 ) Pub Date : 2024-07-09 , DOI: 10.1021/acs.est.4c02769 Xiaoyu Shan 1 , Philip A Wasson 1, 2 , YuanQiao Rao 3 , Scott Backer 4 , Lyndsay Leal 4 , Vurtice C Albright 5 , Wei Gao 3 , Yunzhou Chai 5 , Andreas Sichert 1 , Shaul Pollak 1 , Ligeng Yin 4 , Otto X Cordero 1
Environmental Science & Technology ( IF 10.8 ) Pub Date : 2024-07-09 , DOI: 10.1021/acs.est.4c02769 Xiaoyu Shan 1 , Philip A Wasson 1, 2 , YuanQiao Rao 3 , Scott Backer 4 , Lyndsay Leal 4 , Vurtice C Albright 5 , Wei Gao 3 , Yunzhou Chai 5 , Andreas Sichert 1 , Shaul Pollak 1 , Ligeng Yin 4 , Otto X Cordero 1
Affiliation
|
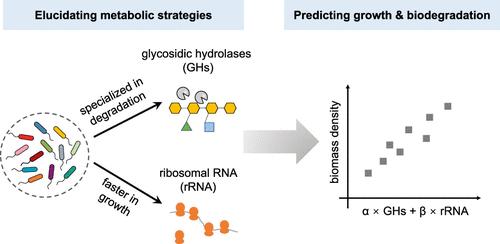
Natural and chemically modified polysaccharides are extensively employed across a wide array of industries, leading to their prevalence in the waste streams of industrialized societies. With projected increasing demand, a pressing challenge is to swiftly assess and predict their biodegradability to inform the development of new sustainable materials. In this study, we developed a scalable method to evaluate polysaccharide breakdown by measuring microbial growth and analyzing microbial genomes. Our approach, applied to polysaccharides with various structures, correlates strongly with well-established regulatory methods based on oxygen demand. We show that modifications to the polysaccharide structure decreased degradability and favored the growth of microbes adapted to break down chemically modified sugars. More broadly, we discovered two main types of microbial communities associated with different polysaccharide structures─one dominated by fast-growing microbes and another by specialized degraders. Surprisingly, we were able to predict biodegradation rates based only on two genomic features that define these communities: the abundance of genes related to rRNA (indicating fast growth) and the abundance of glycoside hydrolases (enzymes that break down polysaccharides), which together predict nearly 70% of the variation in polysaccharide breakdown. This suggests a trade-off, whereby microbes are either adapted for fast growth or for degrading complex polysaccharide chains, but not both. Finally, we observe that viral elements (prophages) encoded in the genomes of degrading microbes are induced in easily degradable polysaccharides, leading to complex dynamics in biomass accumulation during degradation. In summary, our work provides a practical approach for efficiently assessing polymer degradability and offers genomic insights into how microbes break down polysaccharides.
中文翻译:

简单的基因组特征预测多糖生物降解率
天然和化学改性的多糖广泛应用于各个行业,导致它们在工业化社会的废物流中普遍存在。随着预计需求的不断增加,一个紧迫的挑战是迅速评估和预测其生物降解性,为新型可持续材料的开发提供信息。在这项研究中,我们开发了一种可扩展的方法,通过测量微生物生长和分析微生物基因组来评估多糖分解。我们的方法适用于具有各种结构的多糖,与基于需氧量的成熟调节方法密切相关。我们表明,对多糖结构的修饰降低了可降解性,并有利于适合分解化学修饰糖的微生物的生长。更广泛地说,我们发现了与不同多糖结构相关的两种主要微生物群落——一种以快速生长的微生物为主,另一种以专门的降解者为主。令人惊讶的是,我们能够仅根据定义这些群落的两个基因组特征来预测生物降解率:与 rRNA 相关的基因的丰度(表明快速生长)和糖苷水解酶(分解多糖的酶)的丰度,它们一起预测近70% 的多糖分解变化。这表明了一种权衡,即微生物要么适应快速生长,要么适应降解复杂的多糖链,但不能两者兼而有之。最后,我们观察到,降解微生物基因组中编码的病毒元件(原噬菌体)在易于降解的多糖中被诱导,导致降解过程中生物量积累的复杂动态。 总之,我们的工作提供了一种有效评估聚合物降解性的实用方法,并提供了关于微生物如何分解多糖的基因组见解。
更新日期:2024-07-09
中文翻译:

简单的基因组特征预测多糖生物降解率
天然和化学改性的多糖广泛应用于各个行业,导致它们在工业化社会的废物流中普遍存在。随着预计需求的不断增加,一个紧迫的挑战是迅速评估和预测其生物降解性,为新型可持续材料的开发提供信息。在这项研究中,我们开发了一种可扩展的方法,通过测量微生物生长和分析微生物基因组来评估多糖分解。我们的方法适用于具有各种结构的多糖,与基于需氧量的成熟调节方法密切相关。我们表明,对多糖结构的修饰降低了可降解性,并有利于适合分解化学修饰糖的微生物的生长。更广泛地说,我们发现了与不同多糖结构相关的两种主要微生物群落——一种以快速生长的微生物为主,另一种以专门的降解者为主。令人惊讶的是,我们能够仅根据定义这些群落的两个基因组特征来预测生物降解率:与 rRNA 相关的基因的丰度(表明快速生长)和糖苷水解酶(分解多糖的酶)的丰度,它们一起预测近70% 的多糖分解变化。这表明了一种权衡,即微生物要么适应快速生长,要么适应降解复杂的多糖链,但不能两者兼而有之。最后,我们观察到,降解微生物基因组中编码的病毒元件(原噬菌体)在易于降解的多糖中被诱导,导致降解过程中生物量积累的复杂动态。 总之,我们的工作提供了一种有效评估聚合物降解性的实用方法,并提供了关于微生物如何分解多糖的基因组见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号