当前位置:
X-MOL 学术
›
Biotechnol. Adv.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Machine learning for the advancement of genome-scale metabolic modeling
Biotechnology Advances ( IF 12.1 ) Pub Date : 2024-06-27 , DOI: 10.1016/j.biotechadv.2024.108400 Pritam Kundu 1 , Satyajit Beura 2 , Suman Mondal 3 , Amit Kumar Das 2 , Amit Ghosh 4
Biotechnology Advances ( IF 12.1 ) Pub Date : 2024-06-27 , DOI: 10.1016/j.biotechadv.2024.108400 Pritam Kundu 1 , Satyajit Beura 2 , Suman Mondal 3 , Amit Kumar Das 2 , Amit Ghosh 4
Affiliation
|
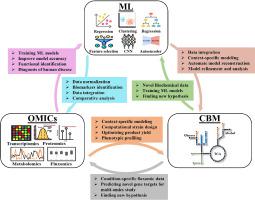
Constraint-based modeling (CBM) has evolved as the core systems biology tool to map the interrelations between genotype, phenotype, and external environment. The recent advancement of high-throughput experimental approaches and multi-omics strategies has generated a plethora of new and precise information from wide-ranging biological domains. On the other hand, the continuously growing field of machine learning (ML) and its specialized branch of deep learning (DL) provide essential computational architectures for decoding complex and heterogeneous biological data. In recent years, both multi-omics and ML have assisted in the escalation of CBM. Condition-specific omics data, such as transcriptomics and proteomics, helped contextualize the model prediction while analyzing a particular phenotypic signature. At the same time, the advanced ML tools have eased the model reconstruction and analysis to increase the accuracy and prediction power. However, the development of these multi-disciplinary methodological frameworks mainly occurs independently, which limits the concatenation of biological knowledge from different domains. Hence, we have reviewed the potential of integrating multi-disciplinary tools and strategies from various fields, such as synthetic biology, CBM, omics, and ML, to explore the biochemical phenomenon beyond the conventional biological dogma. How the integrative knowledge of these intersected domains has improved bioengineering and biomedical applications has also been highlighted. We categorically explained the conventional genome-scale metabolic model (GEM) reconstruction tools and their improvement strategies through ML paradigms. Further, the crucial role of ML and DL in omics data restructuring for GEM development has also been briefly discussed. Finally, the case-study-based assessment of the state-of-the-art method for improving biomedical and metabolic engineering strategies has been elaborated. Therefore, this review demonstrates how integrating experimental and in silico strategies can help map the ever-expanding knowledge of biological systems driven by condition-specific cellular information. This multiview approach will elevate the application of ML-based CBM in the biomedical and bioengineering fields for the betterment of society and the environment.
中文翻译:

机器学习促进基因组规模代谢建模的进步
基于约束的建模 (CBM) 已发展成为绘制基因型、表型和外部环境之间相互关系的核心系统生物学工具。高通量实验方法和多组学策略的最新进展已经从广泛的生物领域产生了大量新的、精确的信息。另一方面,不断发展的机器学习(ML)领域及其专业分支深度学习(DL)为解码复杂和异构的生物数据提供了必要的计算架构。近年来,多组学和机器学习都有助于 CBM 的升级。特定条件的组学数据(例如转录组学和蛋白质组学)有助于在分析特定表型特征时将模型预测置于背景中。同时,先进的机器学习工具简化了模型重建和分析,提高了准确性和预测能力。然而,这些多学科方法论框架的发展主要是独立发生的,这限制了不同领域的生物学知识的串联。因此,我们回顾了整合合成生物学、CBM、组学和机器学习等各个领域的多学科工具和策略来探索传统生物学教条之外的生化现象的潜力。这些交叉领域的综合知识如何改进生物工程和生物医学应用也得到了强调。我们通过机器学习范式明确解释了传统的基因组规模代谢模型(GEM)重建工具及其改进策略。此外,还简要讨论了 ML 和 DL 在 GEM 开发的组学数据重组中的关键作用。 最后,阐述了基于案例研究的对改进生物医学和代谢工程策略的最先进方法的评估。因此,这篇综述展示了如何整合实验和计算机策略来帮助绘制由特定条件的细胞信息驱动的生物系统不断扩展的知识。这种多视图方法将提升基于机器学习的 CBM 在生物医学和生物工程领域的应用,以改善社会和环境。
更新日期:2024-06-27
中文翻译:

机器学习促进基因组规模代谢建模的进步
基于约束的建模 (CBM) 已发展成为绘制基因型、表型和外部环境之间相互关系的核心系统生物学工具。高通量实验方法和多组学策略的最新进展已经从广泛的生物领域产生了大量新的、精确的信息。另一方面,不断发展的机器学习(ML)领域及其专业分支深度学习(DL)为解码复杂和异构的生物数据提供了必要的计算架构。近年来,多组学和机器学习都有助于 CBM 的升级。特定条件的组学数据(例如转录组学和蛋白质组学)有助于在分析特定表型特征时将模型预测置于背景中。同时,先进的机器学习工具简化了模型重建和分析,提高了准确性和预测能力。然而,这些多学科方法论框架的发展主要是独立发生的,这限制了不同领域的生物学知识的串联。因此,我们回顾了整合合成生物学、CBM、组学和机器学习等各个领域的多学科工具和策略来探索传统生物学教条之外的生化现象的潜力。这些交叉领域的综合知识如何改进生物工程和生物医学应用也得到了强调。我们通过机器学习范式明确解释了传统的基因组规模代谢模型(GEM)重建工具及其改进策略。此外,还简要讨论了 ML 和 DL 在 GEM 开发的组学数据重组中的关键作用。 最后,阐述了基于案例研究的对改进生物医学和代谢工程策略的最先进方法的评估。因此,这篇综述展示了如何整合实验和计算机策略来帮助绘制由特定条件的细胞信息驱动的生物系统不断扩展的知识。这种多视图方法将提升基于机器学习的 CBM 在生物医学和生物工程领域的应用,以改善社会和环境。































 京公网安备 11010802027423号
京公网安备 11010802027423号