当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Mutation-Selected Amplification droplet digital PCR: A new single nucleotide variant detection assay for [formula omitted] mutant in tumor and plasma samples
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2024-07-01 , DOI: 10.1016/j.aca.2024.342929 Ling Hu , Yuan-Ye Ji , Peng Zhu , Ren-Quan Lu
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2024-07-01 , DOI: 10.1016/j.aca.2024.342929 Ling Hu , Yuan-Ye Ji , Peng Zhu , Ren-Quan Lu
|
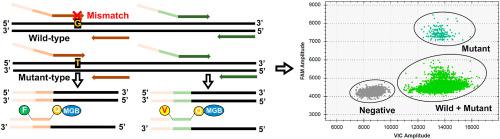
The early detection of gene mutations in physiological and pathological processes is a powerful approach to guide decisions in precision medicine. However, detecting low-copy mutant DNA from clinical samples poses a challenge due to the enrichment of wild-type DNA backgrounds. In this study, we devised a novel strategy, named Mutation-Selected Amplification droplet digital PCR (MSA-ddPCR), to quantitatively analyze single nucleotide variants (SNVs) at low variant allele frequencies (VAFs). Using (a hotspot mutation associated with hepatocellular carcinoma) as a model, we optimized the concentration ratio of primers, the annealing temperature and nucleic acid amplification modifiers. Subsequently, we evaluated the linear range and precision of MSA-ddPCR by detecting and () plasmid DNA, respectively. MSA-ddPCR demonstrated superior ability to discriminate between mutant DNA and wild-type DNA compared to traditional -MGB PCR. We further applied MSA-ddPCR to analyze the mutation in 20 plasma samples and 15 formalin-fixed paraffin-embedded (FFPE) tissue samples, and assessed the agreement rates between MSA-ddPCR and amplicon high-throughput sequencing. The results showed that the limit of blanks of MSA-ddPCR are 0.449 copies μL in the FAM channel and 0.452 copies μL in the VIC channel. MSA-ddPCR could accurately quantify VAFs as low as 0.01 %, surpassing existing PCR and next-generation sequencing (NGS) methods. In the detection of clinical samples, a high correlation was found between MSA-ddPCR and amplicon high-throughput sequencing. Additionally, MSA-ddPCR outperformed sequencing methods in terms of detection time and simplicity of data analysis. MSA-ddPCR can be easily implemented into clinical practice and serve as a robust tool for detecting mutant genes due to its high sensitivity and accuracy.
中文翻译:

突变选择扩增液滴数字 PCR:一种新的单核苷酸变异检测方法,用于检测肿瘤和血浆样本中的[公式省略]突变体
生理和病理过程中基因突变的早期检测是指导精准医学决策的有力方法。然而,由于野生型 DNA 背景的富集,从临床样本中检测低拷贝突变 DNA 提出了挑战。在这项研究中,我们设计了一种名为突变选择扩增液滴数字 PCR (MSA-ddPCR) 的新策略,用于定量分析低变异等位基因频率 (VAF) 下的单核苷酸变异 (SNV)。以(与肝细胞癌相关的热点突变)为模型,我们优化了引物浓度比、退火温度和核酸扩增调节剂。随后,我们分别通过检测 和 () 质粒 DNA 来评估 MSA-ddPCR 的线性范围和精度。与传统的 -MGB PCR 相比,MSA-ddPCR 表现出卓越的区分突变 DNA 和野生型 DNA 的能力。我们进一步应用 MSA-ddPCR 分析了 20 个血浆样本和 15 个福尔马林固定石蜡包埋 (FFPE) 组织样本中的突变,并评估了 MSA-ddPCR 与扩增子高通量测序之间的一致率。结果表明,MSA-ddPCR在FAM通道中的空白限为0.449拷贝μL,在VIC通道中的空白限为0.452拷贝μL。 MSA-ddPCR 可以准确定量低至 0.01% 的 VAF,超越了现有的 PCR 和下一代测序 (NGS) 方法。在临床样本的检测中,发现MSA-ddPCR与扩增子高通量测序之间存在高度相关性。此外,MSA-ddPCR 在检测时间和数据分析简单性方面优于测序方法。 MSA-ddPCR 可以很容易地应用于临床实践,并且由于其高灵敏度和准确性而成为检测突变基因的强大工具。
更新日期:2024-07-01
中文翻译:

突变选择扩增液滴数字 PCR:一种新的单核苷酸变异检测方法,用于检测肿瘤和血浆样本中的[公式省略]突变体
生理和病理过程中基因突变的早期检测是指导精准医学决策的有力方法。然而,由于野生型 DNA 背景的富集,从临床样本中检测低拷贝突变 DNA 提出了挑战。在这项研究中,我们设计了一种名为突变选择扩增液滴数字 PCR (MSA-ddPCR) 的新策略,用于定量分析低变异等位基因频率 (VAF) 下的单核苷酸变异 (SNV)。以(与肝细胞癌相关的热点突变)为模型,我们优化了引物浓度比、退火温度和核酸扩增调节剂。随后,我们分别通过检测 和 () 质粒 DNA 来评估 MSA-ddPCR 的线性范围和精度。与传统的 -MGB PCR 相比,MSA-ddPCR 表现出卓越的区分突变 DNA 和野生型 DNA 的能力。我们进一步应用 MSA-ddPCR 分析了 20 个血浆样本和 15 个福尔马林固定石蜡包埋 (FFPE) 组织样本中的突变,并评估了 MSA-ddPCR 与扩增子高通量测序之间的一致率。结果表明,MSA-ddPCR在FAM通道中的空白限为0.449拷贝μL,在VIC通道中的空白限为0.452拷贝μL。 MSA-ddPCR 可以准确定量低至 0.01% 的 VAF,超越了现有的 PCR 和下一代测序 (NGS) 方法。在临床样本的检测中,发现MSA-ddPCR与扩增子高通量测序之间存在高度相关性。此外,MSA-ddPCR 在检测时间和数据分析简单性方面优于测序方法。 MSA-ddPCR 可以很容易地应用于临床实践,并且由于其高灵敏度和准确性而成为检测突变基因的强大工具。









































 京公网安备 11010802027423号
京公网安备 11010802027423号