当前位置:
X-MOL 学术
›
J. Adv. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Transcriptomic profiling of gastrointestinal tracts in dairy cattle during lactation reveals molecular adaptations for milk synthesis
Journal of Advanced Research ( IF 11.4 ) Pub Date : 2024-06-24 , DOI: 10.1016/j.jare.2024.06.020 Yahui Gao 1 , George E Liu 2 , Li Ma 3 , Lingzhao Fang 4 , Cong-Jun Li 2 , Ransom L Baldwin 2
Journal of Advanced Research ( IF 11.4 ) Pub Date : 2024-06-24 , DOI: 10.1016/j.jare.2024.06.020 Yahui Gao 1 , George E Liu 2 , Li Ma 3 , Lingzhao Fang 4 , Cong-Jun Li 2 , Ransom L Baldwin 2
Affiliation
|
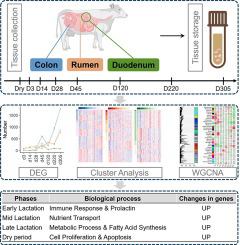
During lactation, dairy cattle’s digestive tract requires significant adaptations to meet the increased nutrient demands for milk production. As we attempt to improve milk-related traits through selective pressure, it is crucial to understand the biological functions of the epithelia of the rumen, small intestine, and colonic tissues in response to changes in physiological state driven by changes in nutrient demands for milk synthesis. In this study, we obtained a total of 108 transcriptome profiles from three tissues (epithelia of the colon, duodenum, and rumen) of five Holstein cows, spanning eight time points from the early, mid, late lactation periods to the dry period. On average 97.06% of reads were successfully mapped to the reference genome assembly ARS-UCD1.2. We analyzed 27,607 gene expression patterns at multiple periods, enabling direct comparisons within and among tissues during different lactation stages, including early and peak lactation. We identified 1645, 813, and 2187 stage-specific genes in the colon, duodenum, and rumen, respectively, which were enriched for common or specific biological functions among different tissues. Time series analysis categorized the expressed genes within each tissue into four clusters. Furthermore, when the three tissues were analyzed collectively, 36 clusters of similarly expressed genes were identified. By integrating other comprehensive approaches such as gene co-expression analyses, functional enrichment, and cell type deconvolution, we gained profound insights into cattle lactation, revealing tissue-specific characteristics of the gastrointestinal tract and shedding light on the intricate molecular adaptations involved in nutrient absorption, immune regulation, and cellular processes for milk synthesis during lactation.
中文翻译:

哺乳期奶牛胃肠道的转录组分析揭示了牛奶合成的分子适应
在哺乳期间,奶牛的消化道需要进行显着的适应,以满足产奶时增加的营养需求。当我们试图通过选择压力改善与乳相关的性状时,了解瘤胃、小肠和结肠组织上皮细胞响应乳合成营养需求变化所驱动的生理状态变化的生物学功能至关重要。在本研究中,我们从5头荷斯坦奶牛的三个组织(结肠上皮、十二指肠和瘤胃)中获得了总共108个转录组图谱,跨越泌乳期早期、中期、晚期到干奶期的8个时间点。平均 97.06% 的读数成功映射到参考基因组组装 ARS-UCD1.2。我们分析了多个时期的 27,607 个基因表达模式,从而能够在不同泌乳阶段(包括泌乳早期和高峰期)的组织内部和组织之间进行直接比较。我们在结肠、十二指肠和瘤胃中分别鉴定了 1645、813 和 2187 个阶段特异性基因,这些基因在不同组织之间具有共同或特定的生物学功能。时间序列分析将每个组织内的表达基因分为四类。此外,当对这三种组织进行共同分析时,发现了 36 个表达相似的基因簇。 通过整合基因共表达分析、功能富集和细胞类型解卷积等其他综合方法,我们对牛泌乳期有了深刻的了解,揭示了胃肠道的组织特异性特征,并揭示了营养吸收中涉及的复杂分子适应、免疫调节和哺乳期间乳汁合成的细胞过程。
更新日期:2024-06-24
中文翻译:

哺乳期奶牛胃肠道的转录组分析揭示了牛奶合成的分子适应
在哺乳期间,奶牛的消化道需要进行显着的适应,以满足产奶时增加的营养需求。当我们试图通过选择压力改善与乳相关的性状时,了解瘤胃、小肠和结肠组织上皮细胞响应乳合成营养需求变化所驱动的生理状态变化的生物学功能至关重要。在本研究中,我们从5头荷斯坦奶牛的三个组织(结肠上皮、十二指肠和瘤胃)中获得了总共108个转录组图谱,跨越泌乳期早期、中期、晚期到干奶期的8个时间点。平均 97.06% 的读数成功映射到参考基因组组装 ARS-UCD1.2。我们分析了多个时期的 27,607 个基因表达模式,从而能够在不同泌乳阶段(包括泌乳早期和高峰期)的组织内部和组织之间进行直接比较。我们在结肠、十二指肠和瘤胃中分别鉴定了 1645、813 和 2187 个阶段特异性基因,这些基因在不同组织之间具有共同或特定的生物学功能。时间序列分析将每个组织内的表达基因分为四类。此外,当对这三种组织进行共同分析时,发现了 36 个表达相似的基因簇。 通过整合基因共表达分析、功能富集和细胞类型解卷积等其他综合方法,我们对牛泌乳期有了深刻的了解,揭示了胃肠道的组织特异性特征,并揭示了营养吸收中涉及的复杂分子适应、免疫调节和哺乳期间乳汁合成的细胞过程。
















































 京公网安备 11010802027423号
京公网安备 11010802027423号