当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Exploring the chemical subspace of RPLC: A data driven approach
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2024-06-20 , DOI: 10.1016/j.aca.2024.342869 Denice van Herwerden , Alexandros Nikolopoulos , Leon P. Barron , Jake W. O’Brien , Bob W.J. Pirok , Kevin V. Thomas , Saer Samanipour
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2024-06-20 , DOI: 10.1016/j.aca.2024.342869 Denice van Herwerden , Alexandros Nikolopoulos , Leon P. Barron , Jake W. O’Brien , Bob W.J. Pirok , Kevin V. Thomas , Saer Samanipour
|
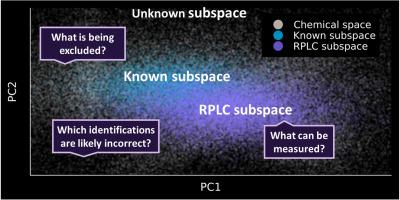
The chemical space is comprised of a vast number of possible structures, of which an unknown portion comprises the human and environmental exposome. Such samples are frequently analyzed using non-targeted analysis via liquid chromatography (LC) coupled to high-resolution mass spectrometry often employing a reversed phase (RP) column. However, prior to analysis, the contents of these samples are unknown and could be comprised of thousands of known and unknown chemical constituents. Moreover, it is unknown which part of the chemical space is sufficiently retained and eluted using RPLC. We present a generic framework that uses a data driven approach to predict whether molecules fall ‘inside’, ‘maybe’ inside, or ‘outside’ of the RPLC subspace. Firstly, three retention index random forest (RF) regression models were constructed that showed that molecular fingerprints are able to predict RPLC retention behavior. Secondly, these models were used to set up the dataset for building an RPLC RF classification model. The RPLC classification model was able to correctly predict whether a chemical belonged to the RPLC subspace with an accuracy of 92% for the testing set. Finally, applying this model to the 91 737 small molecules (i.e., ≤1 000 Da) in NORMAN SusDat showed that 19.1% fall ‘outside’ of the RPLC subspace. The RPLC chemical space model provides a major step towards mapping the chemical space and is able to assess whether chemicals can potentially be measured with an RPLC method (i.e., not every RPLC method) or if a different selectivity should be considered. Moreover, knowing which chemicals are outside of the RPLC subspace can assist in reducing potential candidates for library searching and avoid screening for chemicals that will not be present in RPLC data.
中文翻译:

探索 RPLC 的化学子空间:数据驱动方法
化学空间由大量可能的结构组成,其中未知的部分包括人类和环境暴露组。此类样品经常通过液相色谱 (LC) 与通常采用反相 (RP) 柱的高分辨率质谱联用进行非靶向分析。然而,在分析之前,这些样品的内容是未知的,并且可能由数千种已知和未知的化学成分组成。此外,尚不清楚化学空间的哪一部分被 RPLC 充分保留和洗脱。我们提出了一个通用框架,使用数据驱动的方法来预测分子是否落在 RPLC 子空间的“内部”、“可能”内部或“外部”。首先,构建了三个保留指数随机森林 (RF) 回归模型,表明分子指纹能够预测 RPLC 保留行为。其次,这些模型用于建立用于构建 RPLC RF 分类模型的数据集。 RPLC 分类模型能够正确预测某种化学品是否属于 RPLC 子空间,测试集的准确率为 92%。最后,将该模型应用于 NORMAN SusDat 中的 91 737 个小分子(即 ≤ 1 000 Da),结果表明 19.1% 落在 RPLC 子空间“外部”。 RPLC 化学空间模型为绘制化学空间迈出了重要一步,并且能够评估化学品是否可以使用 RPLC 方法(即并非每种 RPLC 方法)进行测量,或者是否应考虑不同的选择性。此外,了解哪些化学物质在 RPLC 子空间之外可以帮助减少库搜索的潜在候选物质,并避免筛选 RPLC 数据中不存在的化学物质。
更新日期:2024-06-20
中文翻译:

探索 RPLC 的化学子空间:数据驱动方法
化学空间由大量可能的结构组成,其中未知的部分包括人类和环境暴露组。此类样品经常通过液相色谱 (LC) 与通常采用反相 (RP) 柱的高分辨率质谱联用进行非靶向分析。然而,在分析之前,这些样品的内容是未知的,并且可能由数千种已知和未知的化学成分组成。此外,尚不清楚化学空间的哪一部分被 RPLC 充分保留和洗脱。我们提出了一个通用框架,使用数据驱动的方法来预测分子是否落在 RPLC 子空间的“内部”、“可能”内部或“外部”。首先,构建了三个保留指数随机森林 (RF) 回归模型,表明分子指纹能够预测 RPLC 保留行为。其次,这些模型用于建立用于构建 RPLC RF 分类模型的数据集。 RPLC 分类模型能够正确预测某种化学品是否属于 RPLC 子空间,测试集的准确率为 92%。最后,将该模型应用于 NORMAN SusDat 中的 91 737 个小分子(即 ≤ 1 000 Da),结果表明 19.1% 落在 RPLC 子空间“外部”。 RPLC 化学空间模型为绘制化学空间迈出了重要一步,并且能够评估化学品是否可以使用 RPLC 方法(即并非每种 RPLC 方法)进行测量,或者是否应考虑不同的选择性。此外,了解哪些化学物质在 RPLC 子空间之外可以帮助减少库搜索的潜在候选物质,并避免筛选 RPLC 数据中不存在的化学物质。











































 京公网安备 11010802027423号
京公网安备 11010802027423号