当前位置:
X-MOL 学术
›
Metab. Eng.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A long-term growth stable Halomonas sp. deleted with multiple transposases guided by its metabolic network model Halo-ecGEM
Metabolic Engineering ( IF 6.8 ) Pub Date : 2024-06-18 , DOI: 10.1016/j.ymben.2024.06.004 Lizhan Zhang 1 , Jian-Wen Ye 1 , Gang Li 2 , Helen Park 1 , Hao Luo 2 , Yina Lin 1 , Shaowei Li 1 , Weinan Yang 1 , Yuying Guan 1 , Fuqing Wu 3 , Wuzhe Huang 4 , Qiong Wu 5 , Nigel S Scrutton 6 , Jens Nielsen 7 , Guo-Qiang Chen 8
Metabolic Engineering ( IF 6.8 ) Pub Date : 2024-06-18 , DOI: 10.1016/j.ymben.2024.06.004 Lizhan Zhang 1 , Jian-Wen Ye 1 , Gang Li 2 , Helen Park 1 , Hao Luo 2 , Yina Lin 1 , Shaowei Li 1 , Weinan Yang 1 , Yuying Guan 1 , Fuqing Wu 3 , Wuzhe Huang 4 , Qiong Wu 5 , Nigel S Scrutton 6 , Jens Nielsen 7 , Guo-Qiang Chen 8
Affiliation
|
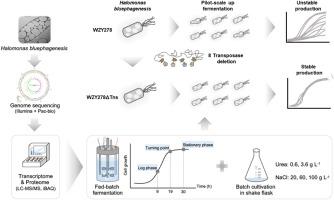
Microbial instability is a common problem during bio-production based on microbial hosts. has been developed as a chassis for next generation industrial biotechnology (NGIB) under open and unsterile conditions. However, the hidden genomic information and peculiar metabolism have significantly hampered its deep exploitation for cell-factory engineering. Based on the freshly completed genome sequence of TD01, which reveals 1889 biological process-associated genes grouped into 84 GO-slim terms. An enzyme constrained genome-scale metabolic model Halo-ecGEM was constructed, which showed strong ability to simulate fed-batch fermentations. A visible salt-stress responsive landscape was achieved by combining GO-slim term enrichment and CVT-based omics profiling, demonstrating that cells deploy most of the protein resources by force to support the essential activity of translation and protein metabolism when exposed to salt stress. Under the guidance of Halo-ecGEM, eight transposases were deleted, leading to a significantly enhanced stability for its growth and bioproduction of various polyhydroxyalkanoates (PHA) including 3-hydroxybutyrate (3HB) homopolymer PHB, 3HB and 3-hydroxyvalerate (3HV) copolymer PHBV, as well as 3HB and 4-hydroxyvalerate (4HB) copolymer P34HB. This study sheds new light on the metabolic characteristics and stress-response landscape of , achieving for the first time to construct a long-term growth stable chassis for industrial applications. For the first time, it was demonstrated that genome encoded transposons are the reason for microbial instability during growth in flasks and fermentors.
中文翻译:

长期生长稳定的盐单胞菌。由其代谢网络模型 Halo-ecGEM 引导的多个转座酶删除
微生物不稳定性是基于微生物宿主的生物生产过程中的常见问题。已开发为开放和非无菌条件下下一代工业生物技术 (NGIB) 的底盘。然而,隐藏的基因组信息和特殊的代谢极大地阻碍了其在细胞工厂工程中的深入开发。基于最新完成的 TD01 基因组序列,该序列揭示了 1889 个生物过程相关基因,分为 84 个 GO-slim 术语。构建了酶约束的基因组规模代谢模型Halo-ecGEM,该模型显示出很强的模拟补料分批发酵的能力。通过结合 GO-slim term 富集和基于 CVT 的组学分析,实现了可见的盐胁迫响应景观,证明细胞在暴露于盐胁迫时强制部署大部分蛋白质资源来支持翻译和蛋白质代谢的基本活动。在Halo-ecGEM的指导下,8个转座酶被删除,导致其生长和生物生产各种聚羟基脂肪酸酯(PHA)的稳定性显着增强,包括3-羟基丁酸酯(3HB)均聚物PHB、3HB和3-羟基戊酸酯(3HV)共聚物PHBV ,以及 3HB 和 4-羟基戊酸酯 (4HB) 共聚物 P34HB。这项研究为了解其代谢特征和应激反应景观提供了新的线索,首次实现了构建适合工业应用的长期生长稳定的底盘。首次证明基因组编码的转座子是烧瓶和发酵罐中微生物生长过程中不稳定的原因。
更新日期:2024-06-18
中文翻译:

长期生长稳定的盐单胞菌。由其代谢网络模型 Halo-ecGEM 引导的多个转座酶删除
微生物不稳定性是基于微生物宿主的生物生产过程中的常见问题。已开发为开放和非无菌条件下下一代工业生物技术 (NGIB) 的底盘。然而,隐藏的基因组信息和特殊的代谢极大地阻碍了其在细胞工厂工程中的深入开发。基于最新完成的 TD01 基因组序列,该序列揭示了 1889 个生物过程相关基因,分为 84 个 GO-slim 术语。构建了酶约束的基因组规模代谢模型Halo-ecGEM,该模型显示出很强的模拟补料分批发酵的能力。通过结合 GO-slim term 富集和基于 CVT 的组学分析,实现了可见的盐胁迫响应景观,证明细胞在暴露于盐胁迫时强制部署大部分蛋白质资源来支持翻译和蛋白质代谢的基本活动。在Halo-ecGEM的指导下,8个转座酶被删除,导致其生长和生物生产各种聚羟基脂肪酸酯(PHA)的稳定性显着增强,包括3-羟基丁酸酯(3HB)均聚物PHB、3HB和3-羟基戊酸酯(3HV)共聚物PHBV ,以及 3HB 和 4-羟基戊酸酯 (4HB) 共聚物 P34HB。这项研究为了解其代谢特征和应激反应景观提供了新的线索,首次实现了构建适合工业应用的长期生长稳定的底盘。首次证明基因组编码的转座子是烧瓶和发酵罐中微生物生长过程中不稳定的原因。































 京公网安备 11010802027423号
京公网安备 11010802027423号