当前位置:
X-MOL 学术
›
Environ. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A genome catalogue of mercury‐methylating bacteria and archaea from sediments of a boreal river facing human disturbances
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-06-23 , DOI: 10.1111/1462-2920.16669 Charlène Lawruk-Desjardins 1 , Veronika Storck 1, 2 , Dominic E Ponton 2 , Marc Amyot 2 , David A Walsh 1
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-06-23 , DOI: 10.1111/1462-2920.16669 Charlène Lawruk-Desjardins 1 , Veronika Storck 1, 2 , Dominic E Ponton 2 , Marc Amyot 2 , David A Walsh 1
Affiliation
|
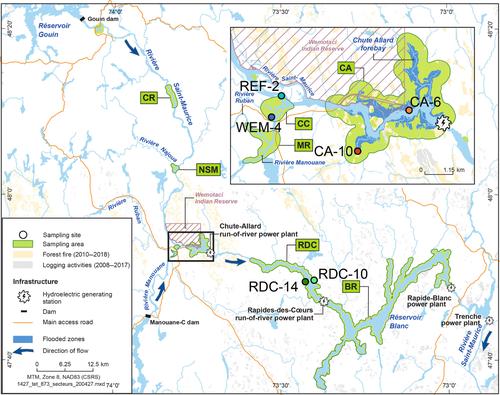
Methyl mercury, a toxic compound, is produced by anaerobic microbes and magnifies in aquatic food webs, affecting the health of animals and humans. The exploration of mercury methylators based on genomes is still limited, especially in the context of river ecosystems. To address this knowledge gap, we developed a genome catalogue of potential mercury‐methylating microorganisms. This was based on the presence of hgcAB from the sediments of a river affected by two run‐of‐river hydroelectric dams, logging activities and a wildfire. Through the use of genome‐resolved metagenomics, we discovered a unique and diverse group of mercury methylators. These were dominated by members of the metabolically versatile Bacteroidota and were particularly rich in microbes that ferment butyrate. By comparing the diversity and abundance of mercury methylators between sites subjected to different disturbances, we found that ongoing disturbances, such as the input of organic matter related to logging activities, were particularly conducive to the establishment of a mercury‐methylating niche. Finally, to gain a deeper understanding of the environmental factors that shape the diversity of mercury methylators, we compared the mercury‐methylating genome catalogue with the broader microbial community. The results suggest that mercury methylators respond to environmental conditions in a manner similar to the overall microbial community. Therefore, it is crucial to interpret the diversity and abundance of mercury methylators within their specific ecological context.
中文翻译:

面临人类干扰的北方河流沉积物中汞甲基化细菌和古细菌的基因组目录
甲基汞是一种有毒化合物,由厌氧微生物产生,并在水生食物网中放大,影响动物和人类的健康。基于基因组的汞甲基化器的探索仍然有限,特别是在河流生态系统的背景下。为了解决这一知识差距,我们开发了潜在汞甲基化微生物的基因组目录。这是基于hgcAB来自受两座径流式水电站大坝、伐木活动和野火影响的河流沉积物。通过使用基因组解析的宏基因组学,我们发现了一组独特且多样化的汞甲基化基因。这些细菌主要由代谢多功能的拟杆菌属成员组成,并且富含发酵丁酸盐的微生物。通过比较遭受不同干扰的地点之间汞甲基化的多样性和丰度,我们发现持续的干扰,例如与伐木活动相关的有机物的输入,特别有利于汞甲基化生态位的建立。最后,为了更深入地了解影响汞甲基化者多样性的环境因素,我们将汞甲基化基因组目录与更广泛的微生物群落进行了比较。结果表明,汞甲基化器对环境条件的反应方式与整个微生物群落相似。因此,在特定的生态背景下解释汞甲基化器的多样性和丰度至关重要。
更新日期:2024-06-23
中文翻译:

面临人类干扰的北方河流沉积物中汞甲基化细菌和古细菌的基因组目录
甲基汞是一种有毒化合物,由厌氧微生物产生,并在水生食物网中放大,影响动物和人类的健康。基于基因组的汞甲基化器的探索仍然有限,特别是在河流生态系统的背景下。为了解决这一知识差距,我们开发了潜在汞甲基化微生物的基因组目录。这是基于hgcAB来自受两座径流式水电站大坝、伐木活动和野火影响的河流沉积物。通过使用基因组解析的宏基因组学,我们发现了一组独特且多样化的汞甲基化基因。这些细菌主要由代谢多功能的拟杆菌属成员组成,并且富含发酵丁酸盐的微生物。通过比较遭受不同干扰的地点之间汞甲基化的多样性和丰度,我们发现持续的干扰,例如与伐木活动相关的有机物的输入,特别有利于汞甲基化生态位的建立。最后,为了更深入地了解影响汞甲基化者多样性的环境因素,我们将汞甲基化基因组目录与更广泛的微生物群落进行了比较。结果表明,汞甲基化器对环境条件的反应方式与整个微生物群落相似。因此,在特定的生态背景下解释汞甲基化器的多样性和丰度至关重要。































 京公网安备 11010802027423号
京公网安备 11010802027423号