当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Native Characterization of Noncanonical Nucleic Acid Thermodynamics via Programmable Dynamic DNA Chemistry
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2024-06-20 , DOI: 10.1021/jacs.4c04721 Yuqin Wu 1 , Guan Alex Wang 1 , Qianfan Yang 1 , Feng Li 1, 2
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2024-06-20 , DOI: 10.1021/jacs.4c04721 Yuqin Wu 1 , Guan Alex Wang 1 , Qianfan Yang 1 , Feng Li 1, 2
Affiliation
|
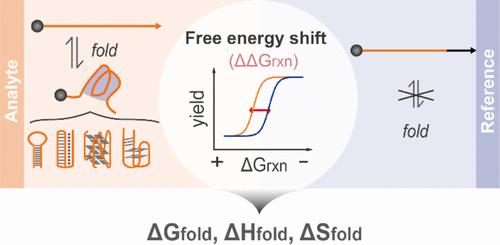
Folding thermodynamics, quantitatively described using parameters such as ΔGfold°, ΔHfold°, and ΔSfold°, is essential for characterizing the stability and functionality of noncanonical nucleic acid structures but remains difficult to measure at the molecular level. Leveraging the programmability of dynamic deoxyribonucleic acid (DNA) chemistry, we introduce a DNA-based molecular tool capable of performing a free energy shift assay (FESA) that directly characterizes the thermodynamics of noncanonical DNA structures in their native environments. FESA operates by the rational design of a reference DNA probe that is energetically equivalent to a target noncanonical nucleic acid structure in a series of toehold-exchange reactions, yet is structurally incapable of folding. As a result, a free energy shift (ΔΔGrxn°) is observed when plotting the reaction yield against the free energy of each toehold-exchange. We mathematically demonstrated that ΔGfold°, ΔHfold°, and ΔSfold° of the analyte can be calculated based on ΔΔGrxn°. After validating FESA using six DNA hairpins by comparing the measured ΔGfold°, ΔHfold°, and ΔSfold° values against predictions made by NUPACK software, we adapted FESA to characterize noncanonical nucleic acid structures, encompassing DNA triplexes, G-quadruplexes, and aptamers. This adaptation enabled the successful characterization of the folding thermodynamics for these complex structures under various experimental conditions. The successful development of FESA marks a paradigm shift and a technical advancement in characterizing the thermodynamics of noncanonical DNA structures through molecular tools. It also opens new avenues for probing fundamental chemical and biophysical questions through the lens of molecular engineering and dynamic DNA chemistry.
中文翻译:

通过可编程动态 DNA 化学对非规范核酸热力学进行本机表征
折叠热力学,使用 ΔG fold ° 、 ΔH fold ° 和 ΔS fold 等参数进行定量描述 ° ,对于表征非规范核酸结构的稳定性和功能性至关重要,但在分子水平上仍然难以测量。利用动态脱氧核糖核酸 (DNA) 化学的可编程性,我们引入了一种基于 DNA 的分子工具,能够执行自由能位移测定 (FESA),直接表征非规范 DNA 结构在其天然环境中的热力学特征。 FESA 通过参考 DNA 探针的合理设计进行操作,该探针在一系列立足点交换反应中与目标非规范核酸结构在能量上等效,但在结构上无法折叠。结果,当将反应产率与每个立足点交换的自由能作图时,观察到自由能偏移(ΔΔG rxn ° )。我们通过数学证明了 ΔG fold ° 、 ΔH fold ° 和 ΔS fold ° 计算分析物的b13>。使用六个 DNA 发夹通过比较测量的 ΔG fold ° 、 ΔH fold ° 和 ΔS fold 验证 FESA 后b20> ° 值与 NUPACK 软件做出的预测相比,我们采用 FESA 来表征非规范核酸结构,包括 DNA 三链体、G-四链体和适体。这种适应性使得能够在各种实验条件下成功表征这些复杂结构的折叠热力学。 FESA 的成功开发标志着通过分子工具表征非规范 DNA 结构热力学的范式转变和技术进步。它还为通过分子工程和动态 DNA 化学的视角探索基本化学和生物物理问题开辟了新途径。
更新日期:2024-06-20
中文翻译:

通过可编程动态 DNA 化学对非规范核酸热力学进行本机表征
折叠热力学,使用 ΔG fold ° 、 ΔH fold ° 和 ΔS fold 等参数进行定量描述 ° ,对于表征非规范核酸结构的稳定性和功能性至关重要,但在分子水平上仍然难以测量。利用动态脱氧核糖核酸 (DNA) 化学的可编程性,我们引入了一种基于 DNA 的分子工具,能够执行自由能位移测定 (FESA),直接表征非规范 DNA 结构在其天然环境中的热力学特征。 FESA 通过参考 DNA 探针的合理设计进行操作,该探针在一系列立足点交换反应中与目标非规范核酸结构在能量上等效,但在结构上无法折叠。结果,当将反应产率与每个立足点交换的自由能作图时,观察到自由能偏移(ΔΔG rxn ° )。我们通过数学证明了 ΔG fold ° 、 ΔH fold ° 和 ΔS fold ° 计算分析物的b13>。使用六个 DNA 发夹通过比较测量的 ΔG fold ° 、 ΔH fold ° 和 ΔS fold 验证 FESA 后b20> ° 值与 NUPACK 软件做出的预测相比,我们采用 FESA 来表征非规范核酸结构,包括 DNA 三链体、G-四链体和适体。这种适应性使得能够在各种实验条件下成功表征这些复杂结构的折叠热力学。 FESA 的成功开发标志着通过分子工具表征非规范 DNA 结构热力学的范式转变和技术进步。它还为通过分子工程和动态 DNA 化学的视角探索基本化学和生物物理问题开辟了新途径。











































 京公网安备 11010802027423号
京公网安备 11010802027423号