当前位置:
X-MOL 学术
›
J. Agric. Food Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Generation of New β-Conglycinin-Deficient Soybean Lines by Editing the lincRNA lincCG1 Using the CRISPR/Cas9 System
Journal of Agricultural and Food Chemistry ( IF 5.7 ) Pub Date : 2024-06-22 , DOI: 10.1021/acs.jafc.4c02269 Bo Song 1, 2 , Tingting Luo 1 , Yuanhang Fan 1 , Ming Li 3 , Zhendong Qiu 1 , Yusu Tian 1 , Yuzhuo Shang 1 , Chongxuan Ma 1 , Chang Liu 1 , Qingqian Cao 1 , Yuhan Peng 1 , Pengfei Xu 1 , Hari B. Krishnan 4, 5 , Zhenhui Wang 6 , Shuzhen Zhang 1 , Shanshan Liu 1
Journal of Agricultural and Food Chemistry ( IF 5.7 ) Pub Date : 2024-06-22 , DOI: 10.1021/acs.jafc.4c02269 Bo Song 1, 2 , Tingting Luo 1 , Yuanhang Fan 1 , Ming Li 3 , Zhendong Qiu 1 , Yusu Tian 1 , Yuzhuo Shang 1 , Chongxuan Ma 1 , Chang Liu 1 , Qingqian Cao 1 , Yuhan Peng 1 , Pengfei Xu 1 , Hari B. Krishnan 4, 5 , Zhenhui Wang 6 , Shuzhen Zhang 1 , Shanshan Liu 1
Affiliation
|
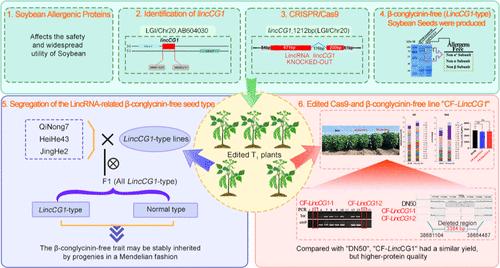
Soybean β-conglycinin is a major allergen that adversely affects the nutritional properties of soybean. Soybean deficient in β-conglycinin is associated with low allergenicity and high nutritional value. Long intergenic noncoding RNAs (lincRNAs) regulate gene expression and are considered important regulators of essential biological processes. Despite increasing knowledge of the functions of lincRNAs, relatively little is known about the effects of lincRNAs on the accumulation of soybean β-conglycinin. The current study presents the identification of a lincRNA lincCG1 that was mapped to the intergenic noncoding region of the β-conglycinin α-subunit locus. The full-length lincCG1 sequence was cloned and found to regulate the expression of soybean seed storage protein (SSP) genes via both cis- and trans-acting regulatory mechanisms. Loss-of-function lincCG1 mutations generated using the clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) system led to the deficiency of the allergenic α′-, α-, and β-subunits of soybean β-conglycinin as well as higher content of proteins, sulfur-containing amino acids, and free arginine. The dominant null allele LincCG1, and consequently, the β-conglycinin-deficient phenotype associated with the lincCG1-gene-edited line was stably inherited by the progenies in a Mendelian fashion. The dominant null allele LincCG1 may therefore be exploited for engineering/developing novel hypoallergenic soybean varieties. Furthermore, Cas9-free and β-conglycinin-deficient homozygous mutant lines were obtained in the T1 generation. This study is the first to employ the CRISPR/Cas9 technology for editing a lincRNA gene associated with the soybean allergenic protein β-conglycinin. Moreover, this study reveals that lincCG1 plays a crucial role in regulating the expression of the β-conglycinin subunit gene cluster, besides highlighting the efficiency of employing the CRISPR/Cas9 system for modulating lincRNAs, and thereby regulating soybean seed components.
中文翻译:

使用 CRISPR/Cas9 系统编辑 lincRNA lincCG1 生成新的 β-伴大豆球蛋白缺陷型大豆品系
大豆β-伴大豆球蛋白是一种主要过敏原,会对大豆的营养特性产生不利影响。缺乏β-伴大豆球蛋白的大豆具有低过敏性和高营养价值。长基因间非编码 RNA (lincRNA) 调节基因表达,被认为是重要生物过程的重要调节因子。尽管人们对 lincRNA 的功能了解越来越多,但关于 lincRNA 对大豆 β-伴大豆球蛋白积累的影响知之甚少。目前的研究鉴定了 lincRNA lincCG1,该 lincRNA lincCG1 被定位到 β-伴大豆球蛋白 α 亚基基因座的基因间非编码区。克隆了全长 lincCG1 序列,发现它可以通过顺式和反式作用调节机制调节大豆种子储存蛋白 (SSP) 基因的表达。使用成簇规则间隔短回文重复序列/CRISPR相关蛋白9 (CRISPR/Cas9) 系统产生的lincCG1功能丧失突变导致大豆β-的致敏性α′-、α-和β-亚基缺乏。伴大豆球蛋白以及较高含量的蛋白质、含硫氨基酸和游离精氨酸。显性无效等位基因 LincCG1,以及因此与 lincCG1 基因编辑系相关的 β-伴大豆球蛋白缺陷表型,以孟德尔方式由后代稳定遗传。因此,显性无效等位基因 LincCG1 可用于工程/开发新型低过敏性大豆品种。此外,在T 1 代中获得了无Cas9和β-伴大豆球蛋白缺陷的纯合突变系。该研究首次采用CRISPR/Cas9技术编辑与大豆过敏蛋白β-伴大豆球蛋白相关的lincRNA基因。 此外,本研究还揭示了lincCG1在调节β-伴大豆球蛋白亚基基因簇的表达中发挥着至关重要的作用,此外还强调了利用CRISPR/Cas9系统调节lincRNA从而调节大豆种子成分的效率。
更新日期:2024-06-22
中文翻译:

使用 CRISPR/Cas9 系统编辑 lincRNA lincCG1 生成新的 β-伴大豆球蛋白缺陷型大豆品系
大豆β-伴大豆球蛋白是一种主要过敏原,会对大豆的营养特性产生不利影响。缺乏β-伴大豆球蛋白的大豆具有低过敏性和高营养价值。长基因间非编码 RNA (lincRNA) 调节基因表达,被认为是重要生物过程的重要调节因子。尽管人们对 lincRNA 的功能了解越来越多,但关于 lincRNA 对大豆 β-伴大豆球蛋白积累的影响知之甚少。目前的研究鉴定了 lincRNA lincCG1,该 lincRNA lincCG1 被定位到 β-伴大豆球蛋白 α 亚基基因座的基因间非编码区。克隆了全长 lincCG1 序列,发现它可以通过顺式和反式作用调节机制调节大豆种子储存蛋白 (SSP) 基因的表达。使用成簇规则间隔短回文重复序列/CRISPR相关蛋白9 (CRISPR/Cas9) 系统产生的lincCG1功能丧失突变导致大豆β-的致敏性α′-、α-和β-亚基缺乏。伴大豆球蛋白以及较高含量的蛋白质、含硫氨基酸和游离精氨酸。显性无效等位基因 LincCG1,以及因此与 lincCG1 基因编辑系相关的 β-伴大豆球蛋白缺陷表型,以孟德尔方式由后代稳定遗传。因此,显性无效等位基因 LincCG1 可用于工程/开发新型低过敏性大豆品种。此外,在T 1 代中获得了无Cas9和β-伴大豆球蛋白缺陷的纯合突变系。该研究首次采用CRISPR/Cas9技术编辑与大豆过敏蛋白β-伴大豆球蛋白相关的lincRNA基因。 此外,本研究还揭示了lincCG1在调节β-伴大豆球蛋白亚基基因簇的表达中发挥着至关重要的作用,此外还强调了利用CRISPR/Cas9系统调节lincRNA从而调节大豆种子成分的效率。






































 京公网安备 11010802027423号
京公网安备 11010802027423号