当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A programmable sensitive platform for pathogen detection based on CRISPR/Cas12a -hybridization chain reaction-poly T-Cu
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2024-06-18 , DOI: 10.1016/j.aca.2024.342888 Haolin Sun , Xiaoyu Zhang , Hainan Ma , Lina Zhang , Yang Zhang , Ruimeng Sun , Haoran Zheng , Han Wang , Jiayu Guo , Yanqi Liu , Yurou wang , Yanfei Qi
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2024-06-18 , DOI: 10.1016/j.aca.2024.342888 Haolin Sun , Xiaoyu Zhang , Hainan Ma , Lina Zhang , Yang Zhang , Ruimeng Sun , Haoran Zheng , Han Wang , Jiayu Guo , Yanqi Liu , Yurou wang , Yanfei Qi
|
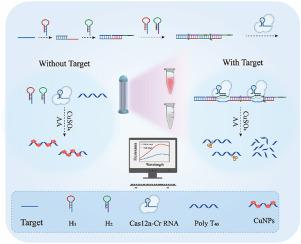
Rapid and sensitive detection of pathogenic bacteria is crucial for disease prevention and control. The CRISPR/Cas12a system with the DNA cleavage capability holds promise in pathogenic bacteria diagnosis. However, the sensitivity of CRISPR-based assays remains a challenge. Herein, we report a versatile and sensitive pathogen sensing platform (HTCas12a) based on the CRISPR/Cas12a system, hybridization chain reaction (HCR) and Poly T-copper fluorescence nanoprobe. The sensitivity is improved by HCR and the Poly-T-Cu reporter probe reduces the overall experiment cost to less than one dollar per sample. Our results demonstrate the specific recognition of target nucleic acid fragments from other pathogens. Furthermore, a good linear correlation between fluorescence intensity and target quantities were achieved with detection limits of 23.36 fM for Target DNA and 4.17 CFU/mL for , respectively. The HTCas12a system offers a universal platform for pathogen detection in various fields, including environmental monitoring, clinical diagnosis, and food safety.
中文翻译:

基于CRISPR/Cas12a-杂交链式反应-聚T-Cu的可编程病原体检测敏感平台
快速、灵敏地检测病原菌对于疾病预防和控制至关重要。具有DNA切割能力的CRISPR/Cas12a系统在病原菌诊断方面具有广阔的前景。然而,基于 CRISPR 的检测的灵敏度仍然是一个挑战。在此,我们报告了一种基于 CRISPR/Cas12a 系统、杂交链式反应 (HCR) 和 Poly T-铜荧光纳米探针的多功能且灵敏的病原体传感平台 (HTCas12a)。 HCR 提高了灵敏度,并且 Poly-T-Cu 报告探针将总体实验成本降低到每个样品不到 1 美元。我们的结果证明了对来自其他病原体的靶核酸片段的特异性识别。此外,荧光强度和目标数量之间实现了良好的线性相关性,目标 DNA 的检测限分别为 23.36 fM,而 4.17 CFU/mL。 HTCas12a系统为环境监测、临床诊断和食品安全等各个领域的病原体检测提供了通用平台。
更新日期:2024-06-18
中文翻译:

基于CRISPR/Cas12a-杂交链式反应-聚T-Cu的可编程病原体检测敏感平台
快速、灵敏地检测病原菌对于疾病预防和控制至关重要。具有DNA切割能力的CRISPR/Cas12a系统在病原菌诊断方面具有广阔的前景。然而,基于 CRISPR 的检测的灵敏度仍然是一个挑战。在此,我们报告了一种基于 CRISPR/Cas12a 系统、杂交链式反应 (HCR) 和 Poly T-铜荧光纳米探针的多功能且灵敏的病原体传感平台 (HTCas12a)。 HCR 提高了灵敏度,并且 Poly-T-Cu 报告探针将总体实验成本降低到每个样品不到 1 美元。我们的结果证明了对来自其他病原体的靶核酸片段的特异性识别。此外,荧光强度和目标数量之间实现了良好的线性相关性,目标 DNA 的检测限分别为 23.36 fM,而 4.17 CFU/mL。 HTCas12a系统为环境监测、临床诊断和食品安全等各个领域的病原体检测提供了通用平台。






































 京公网安备 11010802027423号
京公网安备 11010802027423号