当前位置:
X-MOL 学术
›
Environ. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Multi‐omics analysis of antagonistic interactions among free‐living Pseudonocardia from diverse ecosystems
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-06-20 , DOI: 10.1111/1462-2920.16635 Jonathan Parra 1 , Scott A Jarmusch 2 , Katherine R Duncan 1
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-06-20 , DOI: 10.1111/1462-2920.16635 Jonathan Parra 1 , Scott A Jarmusch 2 , Katherine R Duncan 1
Affiliation
|
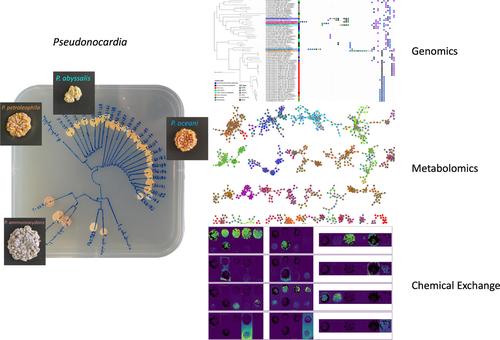
Actinomycetes are a phylogenetically diverse bacterial group which are widely distributed across terrestrial and aquatic ecosystems. Within this order, the genus Pseudonocardia and their specialised metabolites have been the focus of previous ecological studies due to their antagonistic interactions with other microorganisms and their mutualistic interactions with insects. However, the chemical ecology of free‐living Pseudonocardia remains understudied. This study applies a multi‐omics approach to investigate the chemical ecology of free‐living actinomycetes from the genus Pseudonocardia . In a comparative genomics analysis, it was observed that the biosynthetic gene cluster family distribution was influenced mainly by phylogenetic distance rather than the geographic or ecological origin of strains. This finding was also observed in the mass spectrometry‐based metabolomic profiles of nine Pseudonocardia species isolated from marine sediments and two terrestrial species. Antagonist interactions between these 11 species were examined, and matrix‐assisted laser desorption/ionisation‐mass spectrometry imaging was used to examine in situ chemical interactions between the Southern Ocean strains and their phylogenetically close relatives. Overall, it was demonstrated that phylogeny was the main predictor of antagonistic interactions among free‐living Pseudonocardia . Moreover, two features at m /z 441.15 and m /z 332.20 were identified as metabolites related to these interspecies interactions.
中文翻译:

来自不同生态系统的自由生活的假诺卡氏菌之间拮抗相互作用的多组学分析
放线菌是一个系统发育多样化的细菌群,广泛分布在陆地和水生生态系统中。在这个目中,属假诺卡氏菌由于它们与其他微生物的拮抗相互作用以及与昆虫的互利相互作用,它们的特殊代谢物一直是先前生态学研究的焦点。然而,自由生活的化学生态学假诺卡氏菌仍有待研究。本研究应用多组学方法来研究该属自由生活的放线菌的化学生态学假诺卡氏菌。在比较基因组学分析中,观察到生物合成基因簇家族分布主要受系统发育距离的影响,而不是菌株的地理或生态起源的影响。这一发现也在九种基于质谱的代谢组学特征中观察到假诺卡氏菌从海洋沉积物中分离出的物种和两种陆地物种。对这 11 个物种之间的拮抗相互作用进行了检查,并使用基质辅助激光解吸/电离质谱成像来检查南大洋菌株与其系统发育近亲之间的原位化学相互作用。总体而言,系统发育被证明是自由生活的拮抗相互作用的主要预测因素假诺卡氏菌。此外,还有两个特点米/ z 441.15 和米/ z 332.20 被鉴定为与这些种间相互作用相关的代谢物。
更新日期:2024-06-20
中文翻译:

来自不同生态系统的自由生活的假诺卡氏菌之间拮抗相互作用的多组学分析
放线菌是一个系统发育多样化的细菌群,广泛分布在陆地和水生生态系统中。在这个目中,属假诺卡氏菌由于它们与其他微生物的拮抗相互作用以及与昆虫的互利相互作用,它们的特殊代谢物一直是先前生态学研究的焦点。然而,自由生活的化学生态学假诺卡氏菌仍有待研究。本研究应用多组学方法来研究该属自由生活的放线菌的化学生态学假诺卡氏菌。在比较基因组学分析中,观察到生物合成基因簇家族分布主要受系统发育距离的影响,而不是菌株的地理或生态起源的影响。这一发现也在九种基于质谱的代谢组学特征中观察到假诺卡氏菌从海洋沉积物中分离出的物种和两种陆地物种。对这 11 个物种之间的拮抗相互作用进行了检查,并使用基质辅助激光解吸/电离质谱成像来检查南大洋菌株与其系统发育近亲之间的原位化学相互作用。总体而言,系统发育被证明是自由生活的拮抗相互作用的主要预测因素假诺卡氏菌。此外,还有两个特点米/ z 441.15 和米/ z 332.20 被鉴定为与这些种间相互作用相关的代谢物。































 京公网安备 11010802027423号
京公网安备 11010802027423号