当前位置:
X-MOL 学术
›
J. Adv. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Systematic evaluation of single-cell RNA-seq analyses performance based on long-read sequencing platforms
Journal of Advanced Research ( IF 11.4 ) Pub Date : 2024-05-22 , DOI: 10.1016/j.jare.2024.05.020 Enze Deng 1 , Qingmei Shen 2 , Jingna Zhang 3 , Yaowei Fang 3 , Lei Chang 4 , Guanzheng Luo 5 , Xiaoying Fan 2
Journal of Advanced Research ( IF 11.4 ) Pub Date : 2024-05-22 , DOI: 10.1016/j.jare.2024.05.020 Enze Deng 1 , Qingmei Shen 2 , Jingna Zhang 3 , Yaowei Fang 3 , Lei Chang 4 , Guanzheng Luo 5 , Xiaoying Fan 2
Affiliation
|
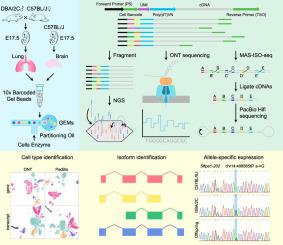
The rapid development of next-generation sequencing (NGS)-based single-cell RNA sequencing (scRNA-seq) allows for detecting and quantifying gene expression in a high-throughput manner, providing a powerful tool for comprehensively understanding cellular function in various biological processes. However, the NGS-based scRNA-seq only quantifies gene expression and cannot reveal the exact transcript structures (isoforms) of each gene due to the limited read length. On the other hand, the long read length of third-generation sequencing (TGS) technologies, including Oxford Nanopore Technologies (ONT) and Pacific Biosciences (PacBio), enable direct reading of intact cDNA molecules. Both ONT and PacBio have been used in conjunction with scRNA-seq, but their performance in single-cell analyses has not been systematically evaluated. To address this, we generated ONT and PacBio data from the same single-cell cDNA libraries containing different amount of cells. Using NGS as a control, we assessed the performance of each platform in cell type identification. Additionally, the reliability in identifying novel isoforms and allele-specific gene/isoform expression by both platforms was verified, providing a systematic evaluation to design the sequencing strategies in single-cell transcriptome studies. Beyond gene expression analysis, which the NGS-based scRNA-seq only affords, TGS-based scRNA-seq achieved gene splicing analyses, identifying novel isoforms. Attribute to higher sequencing quality of PacBio, it outperforms ONT in accuracy of novel transcripts identification and allele-specific gene/isoform expression.
中文翻译:

基于长读长测序平台的单细胞RNA-seq分析性能的系统评估
基于下一代测序(NGS)的单细胞RNA测序(scRNA-seq)的快速发展使得能够以高通量的方式检测和定量基因表达,为全面了解各种生物过程中的细胞功能提供了强大的工具。然而,基于NGS的scRNA-seq仅量化基因表达,由于读长有限,无法揭示每个基因的准确转录物结构(亚型)。另一方面,包括 Oxford Nanopore Technologies (ONT) 和 Pacific Biosciences (PacBio) 在内的第三代测序 (TGS) 技术的长读长能够直接读取完整的 cDNA 分子。 ONT和PacBio都已与scRNA-seq结合使用,但它们在单细胞分析中的性能尚未得到系统评估。为了解决这个问题,我们从包含不同数量细胞的相同单细胞 cDNA 文库中生成了 ONT 和 PacBio 数据。使用 NGS 作为对照,我们评估了每个平台在细胞类型识别方面的性能。此外,两个平台识别新亚型和等位基因特异性基因/亚型表达的可靠性得到了验证,为设计单细胞转录组研究中的测序策略提供了系统评估。除了基于 NGS 的 scRNA-seq 只能提供的基因表达分析之外,基于 TGS 的 scRNA-seq 还实现了基因剪接分析,识别新的亚型。由于 PacBio 具有更高的测序质量,它在新转录本鉴定和等位基因特异性基因/亚型表达的准确性方面优于 ONT。
更新日期:2024-05-22
中文翻译:

基于长读长测序平台的单细胞RNA-seq分析性能的系统评估
基于下一代测序(NGS)的单细胞RNA测序(scRNA-seq)的快速发展使得能够以高通量的方式检测和定量基因表达,为全面了解各种生物过程中的细胞功能提供了强大的工具。然而,基于NGS的scRNA-seq仅量化基因表达,由于读长有限,无法揭示每个基因的准确转录物结构(亚型)。另一方面,包括 Oxford Nanopore Technologies (ONT) 和 Pacific Biosciences (PacBio) 在内的第三代测序 (TGS) 技术的长读长能够直接读取完整的 cDNA 分子。 ONT和PacBio都已与scRNA-seq结合使用,但它们在单细胞分析中的性能尚未得到系统评估。为了解决这个问题,我们从包含不同数量细胞的相同单细胞 cDNA 文库中生成了 ONT 和 PacBio 数据。使用 NGS 作为对照,我们评估了每个平台在细胞类型识别方面的性能。此外,两个平台识别新亚型和等位基因特异性基因/亚型表达的可靠性得到了验证,为设计单细胞转录组研究中的测序策略提供了系统评估。除了基于 NGS 的 scRNA-seq 只能提供的基因表达分析之外,基于 TGS 的 scRNA-seq 还实现了基因剪接分析,识别新的亚型。由于 PacBio 具有更高的测序质量,它在新转录本鉴定和等位基因特异性基因/亚型表达的准确性方面优于 ONT。
















































 京公网安备 11010802027423号
京公网安备 11010802027423号