当前位置:
X-MOL 学术
›
Environ. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Temporal and spatial dynamics within the fungal microbiome of grape fermentation
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-06-01 , DOI: 10.1111/1462-2920.16660 Cristobal A Onetto 1, 2 , Chris M Ward 1 , Steven Van Den Heuvel 1 , Laura Hale 1 , Kathleen Cuijvers 1 , Anthony R Borneman 1, 2
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-06-01 , DOI: 10.1111/1462-2920.16660 Cristobal A Onetto 1, 2 , Chris M Ward 1 , Steven Van Den Heuvel 1 , Laura Hale 1 , Kathleen Cuijvers 1 , Anthony R Borneman 1, 2
Affiliation
|
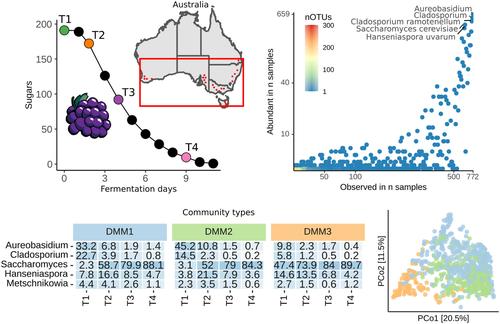
Over 6 years, we conducted an extensive survey of spontaneous grape fermentations, examining 3105 fungal microbiomes across 14 distinct grape‐growing regions. Our investigation into the biodiversity of these fermentations revealed that a small number of highly abundant genera form the core of the initial grape juice microbiome. Consistent with previous studies, we found that the region of origin had the most significant impact on microbial diversity patterns. We also discovered that certain taxa were consistently associated with specific geographical locations and grape varieties, although these taxa represented only a minor portion of the overall diversity in our dataset. Through unsupervised clustering and dimensionality reduction analysis, we identified three unique community types, each exhibiting variations in the abundance of key genera. When we projected these genera onto global branches, it suggested that microbiomes transition between these three broad community types. We further investigated the microbial community composition throughout the fermentation process. Our observations indicated that the initial microbial community composition could predict the diversity during the early stages of fermentation. Notably, Hanseniaspora uvarum emerged as the primary non‐Saccharomyces species within this large collection of samples.
中文翻译:

葡萄发酵真菌微生物组的时空动态
六年多来,我们对葡萄自发发酵进行了广泛的调查,检查了 14 个不同葡萄种植区的 3105 个真菌微生物组。我们对这些发酵的生物多样性的调查表明,少数高度丰富的属构成了最初葡萄汁微生物组的核心。与之前的研究一致,我们发现起源地区对微生物多样性模式的影响最显着。我们还发现某些类群始终与特定的地理位置和葡萄品种相关,尽管这些类群仅占我们数据集中总体多样性的一小部分。通过无监督聚类和降维分析,我们确定了三种独特的群落类型,每种群落类型都表现出关键属丰度的变化。当我们将这些属投影到全球分支时,表明微生物组在这三种广泛的群落类型之间进行转变。我们进一步研究了整个发酵过程中的微生物群落组成。我们的观察表明,初始微生物群落组成可以预测发酵早期阶段的多样性。尤其,葡萄汉逊酵母成为主要的非酵母属这一大样本集合中的物种。
更新日期:2024-06-01
中文翻译:

葡萄发酵真菌微生物组的时空动态
六年多来,我们对葡萄自发发酵进行了广泛的调查,检查了 14 个不同葡萄种植区的 3105 个真菌微生物组。我们对这些发酵的生物多样性的调查表明,少数高度丰富的属构成了最初葡萄汁微生物组的核心。与之前的研究一致,我们发现起源地区对微生物多样性模式的影响最显着。我们还发现某些类群始终与特定的地理位置和葡萄品种相关,尽管这些类群仅占我们数据集中总体多样性的一小部分。通过无监督聚类和降维分析,我们确定了三种独特的群落类型,每种群落类型都表现出关键属丰度的变化。当我们将这些属投影到全球分支时,表明微生物组在这三种广泛的群落类型之间进行转变。我们进一步研究了整个发酵过程中的微生物群落组成。我们的观察表明,初始微生物群落组成可以预测发酵早期阶段的多样性。尤其,葡萄汉逊酵母成为主要的非酵母属这一大样本集合中的物种。































 京公网安备 11010802027423号
京公网安备 11010802027423号