当前位置:
X-MOL 学术
›
Environ. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Specificities and commonalities of the Planctomycetes plasmidome
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-05-11 , DOI: 10.1111/1462-2920.16638 María Del Mar Quiñonero-Coronel 1 , Damien Paul Devos 2 , M Pilar Garcillán-Barcia 1
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-05-11 , DOI: 10.1111/1462-2920.16638 María Del Mar Quiñonero-Coronel 1 , Damien Paul Devos 2 , M Pilar Garcillán-Barcia 1
Affiliation
|
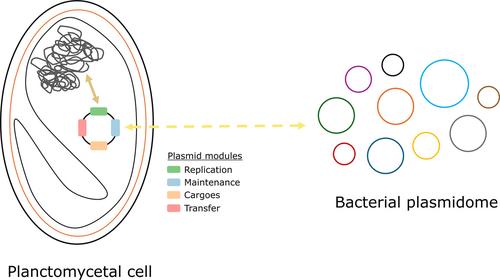
Plasmids, despite their critical role in antibiotic resistance and modern biotechnology, are understood in only a few bacterial groups in terms of their natural ecological dynamics. The bacterial phylum Planctomycetes, known for its unique molecular and cellular biology, has a largely unexplored plasmidome. This study offers a thorough exploration of the diversity of natural plasmids within Planctomycetes, which could serve as a foundation for developing various genetic research tools for this phylum. Planctomycetes plasmids encode a broad range of biological functions and appear to have coevolved significantly with their host chromosomes, sharing many homologues. Recent transfer events of insertion sequences between cohabiting chromosomes and plasmids were also observed. Interestingly, 64% of plasmid genes are distantly related to either chromosomally encoded genes or have homologues in plasmids from other bacterial groups. The planctomycetal plasmidome is composed of 36% exclusive proteins. Most planctomycetal plasmids encode a replication initiation protein from the Replication Protein A family near a putative iteron‐containing replication origin, as well as active type I partition systems. The identification of one conjugative and three mobilizable plasmids suggests the occurrence of horizontal gene transfer via conjugation within this phylum. This comprehensive description enhances our understanding of the plasmidome of Planctomycetes and its potential implications in antibiotic resistance and biotechnology.
中文翻译:

浮霉菌质粒组的特异性和共性
尽管质粒在抗生素耐药性和现代生物技术中发挥着关键作用,但就其自然生态动力学而言,仅在少数细菌群体中被了解。浮霉菌门以其独特的分子和细胞生物学而闻名,其质粒组在很大程度上尚未被探索。这项研究对浮霉菌内天然质粒的多样性进行了彻底的探索,这可以作为开发该门的各种遗传研究工具的基础。浮霉菌质粒编码广泛的生物学功能,并且似乎与其宿主染色体显着共同进化,具有许多同源物。最近还观察到了共居染色体和质粒之间插入序列的转移事件。有趣的是,64% 的质粒基因与染色体编码基因有远缘关系,或者在来自其他细菌群的质粒中具有同源物。浮霉菌质粒组由 36% 的专有蛋白质组成。大多数浮菌质粒编码来自复制蛋白 A 家族的复制起始蛋白,靠近假定的包含 iteron 的复制起点,以及活性 I 型分配系统。一个接合质粒和三个可移动质粒的鉴定表明该门内通过接合发生水平基因转移。这一全面的描述增强了我们对浮霉菌质粒组及其在抗生素耐药性和生物技术方面的潜在影响的理解。
更新日期:2024-05-11
中文翻译:

浮霉菌质粒组的特异性和共性
尽管质粒在抗生素耐药性和现代生物技术中发挥着关键作用,但就其自然生态动力学而言,仅在少数细菌群体中被了解。浮霉菌门以其独特的分子和细胞生物学而闻名,其质粒组在很大程度上尚未被探索。这项研究对浮霉菌内天然质粒的多样性进行了彻底的探索,这可以作为开发该门的各种遗传研究工具的基础。浮霉菌质粒编码广泛的生物学功能,并且似乎与其宿主染色体显着共同进化,具有许多同源物。最近还观察到了共居染色体和质粒之间插入序列的转移事件。有趣的是,64% 的质粒基因与染色体编码基因有远缘关系,或者在来自其他细菌群的质粒中具有同源物。浮霉菌质粒组由 36% 的专有蛋白质组成。大多数浮菌质粒编码来自复制蛋白 A 家族的复制起始蛋白,靠近假定的包含 iteron 的复制起点,以及活性 I 型分配系统。一个接合质粒和三个可移动质粒的鉴定表明该门内通过接合发生水平基因转移。这一全面的描述增强了我们对浮霉菌质粒组及其在抗生素耐药性和生物技术方面的潜在影响的理解。































 京公网安备 11010802027423号
京公网安备 11010802027423号