当前位置:
X-MOL 学术
›
Mol. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Redefining the bacteriophage mv4 site-specific recombination system and the sequence specificity of its attB and core-attP sites
Molecular Microbiology ( IF 2.6 ) Pub Date : 2024-05-05 , DOI: 10.1111/mmi.15275 Kevin Debatisse 1 , Pierre Lopez 1 , Maryse Poli 1 , Philippe Rousseau 2, 3 , Manuel Campos 2, 3 , Michèle Coddeville 2, 3 , Muriel Cocaign-Bousquet 1 , Pascal Le Bourgeois 1, 3
Molecular Microbiology ( IF 2.6 ) Pub Date : 2024-05-05 , DOI: 10.1111/mmi.15275 Kevin Debatisse 1 , Pierre Lopez 1 , Maryse Poli 1 , Philippe Rousseau 2, 3 , Manuel Campos 2, 3 , Michèle Coddeville 2, 3 , Muriel Cocaign-Bousquet 1 , Pascal Le Bourgeois 1, 3
Affiliation
|
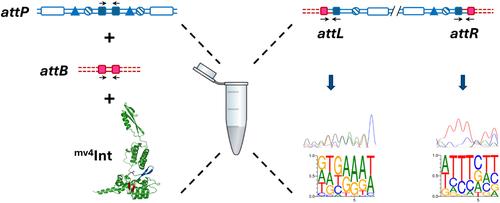
Through their involvement in the integration and excision of a large number of mobile genetic elements, such as phages and integrative and conjugative elements (ICEs), site-specific recombination systems based on heterobivalent tyrosine recombinases play a major role in genome dynamics and evolution. However, despite hundreds of these systems having been identified in genome databases, very few have been described in detail, with none from phages that infect Bacillota (formerly Firmicutes). In this study, we reanalyzed the recombination module of Lactobacillus delbrueckii subsp. bulgaricus phage mv4, previously considered atypical compared with classical systems. Our results reveal that mv4 integrase is a 369 aa protein with all the structural hallmarks of recombinases from the Tn916 family and that it cooperatively interacts with its recombination sites. Using randomized DNA libraries, NGS sequencing, and other molecular approaches, we show that the 21-bp core-attP and attB sites have structural similarities to classical systems only if considering the nucleotide degeneracy, with two 7-bp inverted regions corresponding to mv4Int core-binding sites surrounding a 7-bp strand-exchange region. We also examined the different compositional constraints in the core-binding regions, which define the sequence space of permissible recombination sites.
中文翻译:

重新定义噬菌体mv4位点特异性重组系统及其attB和core-attP位点的序列特异性
基于异二价酪氨酸重组酶的位点特异性重组系统参与大量可移动遗传元件(例如噬菌体以及整合和接合元件(ICE))的整合和切除,在基因组动力学和进化中发挥着重要作用。然而,尽管基因组数据库中已鉴定出数百个此类系统,但详细描述的系统却很少,没有一个来自感染芽孢杆菌(以前称为厚壁菌门)的噬菌体。在本研究中,我们重新分析了德氏乳杆菌亚种的重组模块。保加利亚噬菌体 mv4,以前被认为与经典系统相比是非典型的。我们的结果表明,mv4 整合酶是一种 369 个氨基酸的蛋白质,具有 Tn 916家族重组酶的所有结构特征,并且它与其重组位点协同相互作用。使用随机 DNA 文库、NGS 测序和其他分子方法,我们表明,仅在考虑核苷酸简并性的情况下,21 bp core- attP和attB位点与经典系统具有结构相似性,其中两个 7 bp 反向区域对应于mv4 Int围绕 7 bp 链交换区域的核心结合位点。我们还检查了核心结合区的不同组成限制,这些限制定义了允许重组位点的序列空间。
更新日期:2024-05-05
中文翻译:

重新定义噬菌体mv4位点特异性重组系统及其attB和core-attP位点的序列特异性
基于异二价酪氨酸重组酶的位点特异性重组系统参与大量可移动遗传元件(例如噬菌体以及整合和接合元件(ICE))的整合和切除,在基因组动力学和进化中发挥着重要作用。然而,尽管基因组数据库中已鉴定出数百个此类系统,但详细描述的系统却很少,没有一个来自感染芽孢杆菌(以前称为厚壁菌门)的噬菌体。在本研究中,我们重新分析了德氏乳杆菌亚种的重组模块。保加利亚噬菌体 mv4,以前被认为与经典系统相比是非典型的。我们的结果表明,mv4 整合酶是一种 369 个氨基酸的蛋白质,具有 Tn 916家族重组酶的所有结构特征,并且它与其重组位点协同相互作用。使用随机 DNA 文库、NGS 测序和其他分子方法,我们表明,仅在考虑核苷酸简并性的情况下,21 bp core- attP和attB位点与经典系统具有结构相似性,其中两个 7 bp 反向区域对应于mv4 Int围绕 7 bp 链交换区域的核心结合位点。我们还检查了核心结合区的不同组成限制,这些限制定义了允许重组位点的序列空间。






























 京公网安备 11010802027423号
京公网安备 11010802027423号