Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Tuning the Dynamic Reaction Balance of CRISPR/Cas12a and RPA in One Pot: A Key to Switch Nucleic Acid Quantification
ACS Sensors ( IF 8.2 ) Pub Date : 2024-04-23 , DOI: 10.1021/acssensors.3c02485 Zhihao Yao 1, 2 , Kaiyu He 2 , Hongmei Wang 2 , Suyin Feng 3, 4 , Xiaoqing Ding 5 , Yan Xu 1 , Qiang Wang 2 , Xiahong Xu 2 , Qun Wu 1 , Liu Wang 2, 6
ACS Sensors ( IF 8.2 ) Pub Date : 2024-04-23 , DOI: 10.1021/acssensors.3c02485 Zhihao Yao 1, 2 , Kaiyu He 2 , Hongmei Wang 2 , Suyin Feng 3, 4 , Xiaoqing Ding 5 , Yan Xu 1 , Qiang Wang 2 , Xiahong Xu 2 , Qun Wu 1 , Liu Wang 2, 6
Affiliation
|
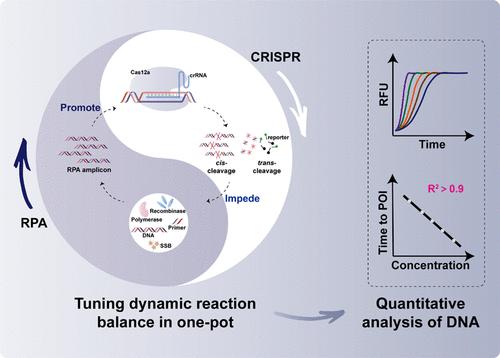
Excavating nucleic acid quantitative capabilities by combining clustered regularly interspaced short palindromic repeats (CRISPR) and isothermal amplification in one pot is of common interest. However, the mutual interference between CRISPR cleavage and isothermal amplification is the primary obstacle to quantitative detection. Though several works have demonstrated enhanced detection sensitivity by reducing the inhibition of CRISPR on amplification in one pot, few paid attention to the amplification process and even dynamic reaction processes between the two. Herein, we find that DNA quantification can be realized by regulating either recombinase polymerase amplification (RPA) efficiency or CRISPR/Cas12a cleaving efficiency (namely, tuning the dynamic reaction balance) in one pot. The sensitive quantification is realized by utilizing dual PAM-free crRNAs for CRISPR/Cas12a recognition. The varied RPA primer concentration with stabilized CRISPR systems significantly affects the amplification efficiency and quantitative performances. Alternatively, quantitative detection can also be achieved by stabilizing the amplification process while regulating the CRISPR/Cas12a concentration. The quantitative capability is proved by detecting DNA targets from Lactobacillus acetotolerans and SARS-CoV-2. The quantitative performance toward real samples is comparable to quantitative real-time PCR for detecting L. acetotolerans spiked in fermented food samples and SARS-CoV-2 clinical samples. We expect that the presented method will be a powerful tool for quantifying other nucleic acid targets.
中文翻译:

一锅调整 CRISPR/Cas12a 和 RPA 的动态反应平衡:切换核酸定量的关键
通过将成簇规则间隔短回文重复序列 (CRISPR) 和等温扩增结合在一锅中来挖掘核酸定量能力是人们普遍感兴趣的。然而,CRISPR裂解和等温扩增之间的相互干扰是定量检测的主要障碍。尽管一些工作已经证明通过减少CRISPR对一锅扩增的抑制来提高检测灵敏度,但很少有人关注扩增过程,甚至两者之间的动态反应过程。在此,我们发现DNA定量可以通过一锅调节重组酶聚合酶扩增(RPA)效率或CRISPR/Cas12a切割效率(即调节动态反应平衡)来实现。利用双不含 PAM 的 crRNA 进行 CRISPR/Cas12a 识别,实现灵敏的定量。稳定的 CRISPR 系统中不同的 RPA 引物浓度会显着影响扩增效率和定量性能。或者,也可以通过稳定扩增过程同时调节CRISPR/Cas12a浓度来实现定量检测。通过检测乙酰耐受乳杆菌和 SARS-CoV-2 的 DNA 靶标证明了定量能力。对真实样品的定量性能与用于检测发酵食品样品和 SARS-CoV-2 临床样品中加标的乙酰耐受乳杆菌的定量实时 PCR 相当。我们期望所提出的方法将成为量化其他核酸靶标的强大工具。
更新日期:2024-04-23
中文翻译:

一锅调整 CRISPR/Cas12a 和 RPA 的动态反应平衡:切换核酸定量的关键
通过将成簇规则间隔短回文重复序列 (CRISPR) 和等温扩增结合在一锅中来挖掘核酸定量能力是人们普遍感兴趣的。然而,CRISPR裂解和等温扩增之间的相互干扰是定量检测的主要障碍。尽管一些工作已经证明通过减少CRISPR对一锅扩增的抑制来提高检测灵敏度,但很少有人关注扩增过程,甚至两者之间的动态反应过程。在此,我们发现DNA定量可以通过一锅调节重组酶聚合酶扩增(RPA)效率或CRISPR/Cas12a切割效率(即调节动态反应平衡)来实现。利用双不含 PAM 的 crRNA 进行 CRISPR/Cas12a 识别,实现灵敏的定量。稳定的 CRISPR 系统中不同的 RPA 引物浓度会显着影响扩增效率和定量性能。或者,也可以通过稳定扩增过程同时调节CRISPR/Cas12a浓度来实现定量检测。通过检测乙酰耐受乳杆菌和 SARS-CoV-2 的 DNA 靶标证明了定量能力。对真实样品的定量性能与用于检测发酵食品样品和 SARS-CoV-2 临床样品中加标的乙酰耐受乳杆菌的定量实时 PCR 相当。我们期望所提出的方法将成为量化其他核酸靶标的强大工具。















































 京公网安备 11010802027423号
京公网安备 11010802027423号