当前位置:
X-MOL 学术
›
Sci. Total Environ.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Isolation and characterization of novel 3,3′-iminodipropionitrile biodegrading Paracoccus communis, from an industrial wastewater treatment bioreactor
Science of the Total Environment ( IF 8.2 ) Pub Date : 2024-04-04 , DOI: 10.1016/j.scitotenv.2024.172099 Sang-Hoon Lee 1 , Na-Kyung Kim 1 , You-Jung Jung 2 , Shin Hae Cho 3 , Onekyun Choi 4 , Jeong-Hoon Lee 1 , Ki-Seung Choi 5 , Hyeokjun Yoon 2 , Moonsuk Hur 2 , Hee-Deung Park 1
Science of the Total Environment ( IF 8.2 ) Pub Date : 2024-04-04 , DOI: 10.1016/j.scitotenv.2024.172099 Sang-Hoon Lee 1 , Na-Kyung Kim 1 , You-Jung Jung 2 , Shin Hae Cho 3 , Onekyun Choi 4 , Jeong-Hoon Lee 1 , Ki-Seung Choi 5 , Hyeokjun Yoon 2 , Moonsuk Hur 2 , Hee-Deung Park 1
Affiliation
|
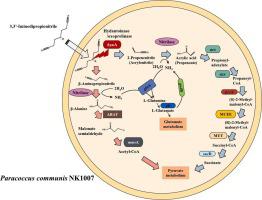
Until now, bacteria able to degrade, 3,3′-iminodipropionitrile (IDPN), a neurotoxin that destroys vestibular hair cells, causing ototoxicity, culminating in irreversible movement disorders, had never been isolated. The aim of this study was to isolate a novel IDPN-biodegrading microorganism and characterize its metabolic pathway. Enrichment was performed by inoculating activated sludge from a wastewater treatment bioreactor that treated IDPN-contaminated wastewater in M9 salt medium, with IDPN as the sole carbon source. A bacterial strain with a spherical morphology that could grow at high concentrations was isolated on a solid medium. Growth of the isolated strain followed the Monod kinetic model. Based on the 16S rRNA gene, the isolate was . Whole-genome sequencing revealed that the isolated possessed the expected full metabolic pathway for IDPN biodegradation. Transcriptome analyses confirmed the overexpression of the gene encoding hydantoinase/oxoprolinase during the exponential growth phase under IDPN-fed conditions, suggesting that the enzyme involved in cleaving the imine bond of IDPN may promote IDPN biodegradation. Additionally, the newly discovered isolate seems to metabolize IDPN through cleavage of the imine bond in IDPN via nitrilase, nitrile hydratase, and amidase reactions. Overall, this study lays the foundation for the application of IDPN-metabolizing bacteria in the remediation of IDPN-contaminated environments.
中文翻译:

从工业废水处理生物反应器中分离和表征新型 3,3'-亚氨基二丙腈生物降解副球菌
迄今为止,尚未分离出能够降解 3,3'-亚氨基二丙腈 (IDPN) 的细菌,这种神经毒素会破坏前庭毛细胞,引起耳毒性,最终导致不可逆的运动障碍。本研究的目的是分离一种新型 IDPN 生物降解微生物并表征其代谢途径。通过接种来自废水处理生物反应器的活性污泥来进行富集,该生物反应器在 M9 盐介质中处理 IDPN 污染的废水,以 IDPN 作为唯一碳源。在固体培养基上分离出一种具有球形形态、可以在高浓度下生长的细菌菌株。分离菌株的生长遵循莫诺动力学模型。根据 16S rRNA 基因,分离株为 .全基因组测序表明,分离物具有预期的 IDPN 生物降解完整代谢途径。转录组分析证实,在 IDPN 喂养条件下,在指数生长期,编码乙内酰脲酶/氧代脯氨酸酶的基因过度表达,表明参与裂解 IDPN 亚胺键的酶可能促进 IDPN 生物降解。此外,新发现的分离物似乎通过腈水解酶、腈水合酶和酰胺酶反应裂解 IDPN 中的亚胺键来代谢 IDPN。总体而言,本研究为IDPN代谢菌在IDPN污染环境修复中的应用奠定了基础。
更新日期:2024-04-04
中文翻译:

从工业废水处理生物反应器中分离和表征新型 3,3'-亚氨基二丙腈生物降解副球菌
迄今为止,尚未分离出能够降解 3,3'-亚氨基二丙腈 (IDPN) 的细菌,这种神经毒素会破坏前庭毛细胞,引起耳毒性,最终导致不可逆的运动障碍。本研究的目的是分离一种新型 IDPN 生物降解微生物并表征其代谢途径。通过接种来自废水处理生物反应器的活性污泥来进行富集,该生物反应器在 M9 盐介质中处理 IDPN 污染的废水,以 IDPN 作为唯一碳源。在固体培养基上分离出一种具有球形形态、可以在高浓度下生长的细菌菌株。分离菌株的生长遵循莫诺动力学模型。根据 16S rRNA 基因,分离株为 .全基因组测序表明,分离物具有预期的 IDPN 生物降解完整代谢途径。转录组分析证实,在 IDPN 喂养条件下,在指数生长期,编码乙内酰脲酶/氧代脯氨酸酶的基因过度表达,表明参与裂解 IDPN 亚胺键的酶可能促进 IDPN 生物降解。此外,新发现的分离物似乎通过腈水解酶、腈水合酶和酰胺酶反应裂解 IDPN 中的亚胺键来代谢 IDPN。总体而言,本研究为IDPN代谢菌在IDPN污染环境修复中的应用奠定了基础。






























 京公网安备 11010802027423号
京公网安备 11010802027423号