当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Enhancing Multi-species Liver Microsomal Stability Prediction through Artificial Intelligence
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-03-18 , DOI: 10.1021/acs.jcim.4c00159 Teng-Zhi Long 1 , De-Jun Jiang 2 , Shao-Hua Shi 3 , You-Chao Deng 1 , Wen-Xuan Wang 1 , Dong-Sheng Cao 1, 3, 4
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-03-18 , DOI: 10.1021/acs.jcim.4c00159 Teng-Zhi Long 1 , De-Jun Jiang 2 , Shao-Hua Shi 3 , You-Chao Deng 1 , Wen-Xuan Wang 1 , Dong-Sheng Cao 1, 3, 4
Affiliation
|
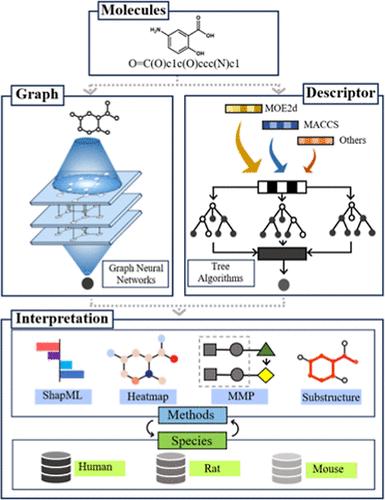
Liver microsomal stability, a crucial aspect of metabolic stability, significantly impacts practical drug discovery. However, current models for predicting liver microsomal stability are based on limited molecular information from a single species. To address this limitation, we constructed the largest public database of compounds from three common species: human, rat, and mouse. Subsequently, we developed a series of classification models using both traditional descriptor-based and classic graph-based machine learning (ML) algorithms. Remarkably, the best-performing models for the three species achieved Matthews correlation coefficients (MCCs) of 0.616, 0.603, and 0.574, respectively, on the test set. Furthermore, through the construction of consensus models based on these individual models, we have demonstrated their superior predictive performance in comparison with the existing models of the same type. To explore the similarities and differences in the properties of liver microsomal stability among multispecies molecules, we conducted preliminary interpretative explorations using the Shapley additive explanations (SHAP) and atom heatmap approaches for the models and misclassified molecules. Additionally, we further investigated representative structural modifications and substructures that decrease the liver microsomal stability in different species using the matched molecule pair analysis (MMPA) method and substructure extraction techniques. The established prediction models, along with insightful interpretation information regarding liver microsomal stability, will significantly contribute to enhancing the efficiency of exploring practical drugs for development.
中文翻译:

通过人工智能增强多物种肝脏微粒体稳定性预测
肝微粒体稳定性是代谢稳定性的一个重要方面,显着影响实际的药物发现。然而,目前预测肝微粒体稳定性的模型是基于来自单一物种的有限分子信息。为了解决这一限制,我们构建了来自三个常见物种(人类、大鼠和小鼠)的最大化合物公共数据库。随后,我们使用传统的基于描述符和经典的基于图的机器学习(ML)算法开发了一系列分类模型。值得注意的是,这三个物种表现最好的模型在测试集上的马修斯相关系数 (MCC) 分别为 0.616、0.603 和 0.574。此外,通过基于这些单独模型构建共识模型,我们证明了它们与现有同类型模型相比具有优越的预测性能。为了探索多物种分子之间肝微粒体稳定性特性的异同,我们使用沙普利附加解释(SHAP)和原子热图方法对模型和错误分类的分子进行了初步解释探索。此外,我们使用匹配分子对分析(MMPA)方法和子结构提取技术进一步研究了降低不同物种肝微粒体稳定性的代表性结构修饰和子结构。建立的预测模型以及有关肝微粒体稳定性的深入解释信息将极大地有助于提高探索实用药物开发的效率。
更新日期:2024-03-18
中文翻译:

通过人工智能增强多物种肝脏微粒体稳定性预测
肝微粒体稳定性是代谢稳定性的一个重要方面,显着影响实际的药物发现。然而,目前预测肝微粒体稳定性的模型是基于来自单一物种的有限分子信息。为了解决这一限制,我们构建了来自三个常见物种(人类、大鼠和小鼠)的最大化合物公共数据库。随后,我们使用传统的基于描述符和经典的基于图的机器学习(ML)算法开发了一系列分类模型。值得注意的是,这三个物种表现最好的模型在测试集上的马修斯相关系数 (MCC) 分别为 0.616、0.603 和 0.574。此外,通过基于这些单独模型构建共识模型,我们证明了它们与现有同类型模型相比具有优越的预测性能。为了探索多物种分子之间肝微粒体稳定性特性的异同,我们使用沙普利附加解释(SHAP)和原子热图方法对模型和错误分类的分子进行了初步解释探索。此外,我们使用匹配分子对分析(MMPA)方法和子结构提取技术进一步研究了降低不同物种肝微粒体稳定性的代表性结构修饰和子结构。建立的预测模型以及有关肝微粒体稳定性的深入解释信息将极大地有助于提高探索实用药物开发的效率。































 京公网安备 11010802027423号
京公网安备 11010802027423号