当前位置:
X-MOL 学术
›
Bioconjugate Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Synthetic and Computational Design Insights toward Mimicking Protein Binding of Phosphate
Bioconjugate Chemistry ( IF 4.0 ) Pub Date : 2024-02-20 , DOI: 10.1021/acs.bioconjchem.3c00454 Whitney C Fowler 1 , Chuting Deng 1 , O Therese Teodoro 1 , Juan J de Pablo 1, 2 , Matthew V Tirrell 1, 2
Bioconjugate Chemistry ( IF 4.0 ) Pub Date : 2024-02-20 , DOI: 10.1021/acs.bioconjchem.3c00454 Whitney C Fowler 1 , Chuting Deng 1 , O Therese Teodoro 1 , Juan J de Pablo 1, 2 , Matthew V Tirrell 1, 2
Affiliation
|
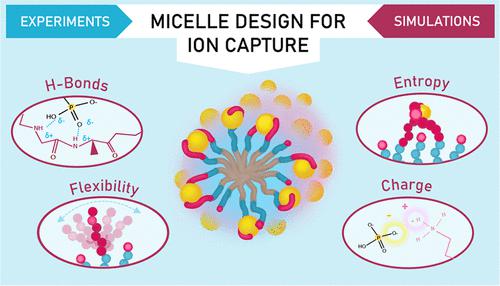
The unique and precise capabilities of proteins are renowned for their specificity and range of application. Effective mimicking of protein-binding offers enticing potential to direct their abilities toward useful applications, but it is nevertheless quite difficult to realize this characteristic of protein behavior in a synthetic material. Here, we design, synthesize, and evaluate experimentally and computationally a series of multicomponent phosphate-binding peptide amphiphile micelles to derive design insights into how protein binding behavior translates to synthetic materials. By inserting the Walker A P-loop binding motif into this peptide synthetic material, we successfully implemented the protein-binding design parameters of hydrogen-bonding and electrostatic interaction to bind phosphate completely and selectively in this highly tunable synthetic platform. Moreover, in this densely arrayed peptide environment, we use molecular dynamics simulations to identify an intriguing mechanistic shift of binding that is inaccessible in traditional proteins, introducing two corresponding new design elements─flexibility and minimization of the loss of entropy due to ion binding, in protein-analogous synthetic materials. We then translate these new design factors to de novo peptide sequences that bind phosphate independent of protein-extracted sequence or conformation. Overall, this work reveals that traditional complex conformational restrictions of binding by proteins can be replaced and repurposed in a multicomponent peptide amphiphile synthetic material, opening up opportunities for future enhanced protein-inspired design.
中文翻译:

模拟磷酸盐蛋白质结合的合成和计算设计见解
蛋白质独特而精确的能力以其特异性和应用范围而闻名。蛋白质结合的有效模拟为将它们的能力引导到有用的应用提供了诱人的潜力,但在合成材料中实现蛋白质行为的这一特征仍然相当困难。在这里,我们设计、合成和实验和计算评估了一系列多组分磷酸盐结合肽两亲性胶束,以获得关于蛋白质结合行为如何转化为合成材料的设计见解。通过将 Walker A P 环结合基序插入这种肽合成材料中,我们成功地实现了氢键和静电相互作用的蛋白质结合设计参数,在这个高度可调的合成平台中完全选择性地结合磷酸盐。此外,在这种密集排列的肽环境中,我们使用分子动力学模拟来识别传统蛋白质中无法实现的有趣的结合机制转变,在蛋白质类似合成材料中引入了两个相应的新设计元素——灵活性和因离子结合而造成的熵损失的最小化。然后,我们将这些新的设计因子转化为结合磷酸盐的从头肽序列,而与蛋白质提取的序列或构象无关。总体而言,这项工作揭示了蛋白质结合的传统复杂构象限制可以在多组分肽两亲性合成材料中被替代和重新利用,为未来增强的蛋白质启发设计开辟了机会。
更新日期:2024-02-20
中文翻译:

模拟磷酸盐蛋白质结合的合成和计算设计见解
蛋白质独特而精确的能力以其特异性和应用范围而闻名。蛋白质结合的有效模拟为将它们的能力引导到有用的应用提供了诱人的潜力,但在合成材料中实现蛋白质行为的这一特征仍然相当困难。在这里,我们设计、合成和实验和计算评估了一系列多组分磷酸盐结合肽两亲性胶束,以获得关于蛋白质结合行为如何转化为合成材料的设计见解。通过将 Walker A P 环结合基序插入这种肽合成材料中,我们成功地实现了氢键和静电相互作用的蛋白质结合设计参数,在这个高度可调的合成平台中完全选择性地结合磷酸盐。此外,在这种密集排列的肽环境中,我们使用分子动力学模拟来识别传统蛋白质中无法实现的有趣的结合机制转变,在蛋白质类似合成材料中引入了两个相应的新设计元素——灵活性和因离子结合而造成的熵损失的最小化。然后,我们将这些新的设计因子转化为结合磷酸盐的从头肽序列,而与蛋白质提取的序列或构象无关。总体而言,这项工作揭示了蛋白质结合的传统复杂构象限制可以在多组分肽两亲性合成材料中被替代和重新利用,为未来增强的蛋白质启发设计开辟了机会。






























 京公网安备 11010802027423号
京公网安备 11010802027423号