Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Programmable Primer Switching for Regulating Enzymatic DNA Circuits
ACS Nano ( IF 15.8 ) Pub Date : 2024-01-29 , DOI: 10.1021/acsnano.3c12000 Yongpeng Zhang 1 , Yiming Chen 2 , Xuan Liu 1 , Qian Ling 2 , Ranfeng Wu 3 , Jing Yang 1 , Cheng Zhang 2
ACS Nano ( IF 15.8 ) Pub Date : 2024-01-29 , DOI: 10.1021/acsnano.3c12000 Yongpeng Zhang 1 , Yiming Chen 2 , Xuan Liu 1 , Qian Ling 2 , Ranfeng Wu 3 , Jing Yang 1 , Cheng Zhang 2
Affiliation
|
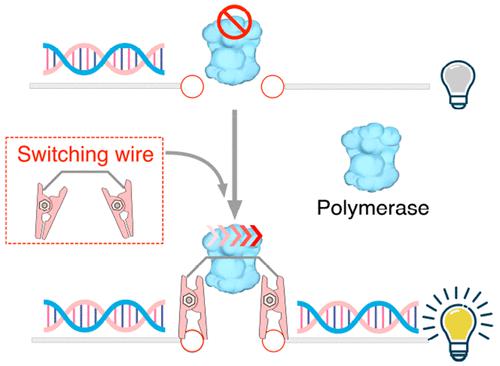
Developing DNA strand displacement reactions (SDRs) offers crucial technical support for regulating artificial nucleic acid circuits and networks. More recently, enzymatic SDR-based DNA circuits have gained significant attention because of their modular design, high orthogonality signaling, and extremely fast reaction rates. Typical enzymatic SDRs are regulated by relatively long primers (20–30 nucleotides) that hybridize to form stable double-stranded structures, facilitating enzyme-initiated events. Implementing more flexible primer-based enzymatic SDR regulations remains challenging due to the lack of convenient and simple primer control mechanism, which consequently limits the development of enzymatic DNA circuits. In this study, we propose an approach, termed primer switching regulation, that implements programmable and flexible regulations of enzymatic circuits by introducing switchable wires into the enzymatic circuits. We applied this method to generate diverse enzymatic DNA circuits, including cascading, fan-in/fan-out, dual-rail, feed-forward, and feedback functions. Through this method, complex circuit functions can be implemented by just introducing additional switching wires without reconstructing the basic circuit frameworks. The method is experimentally demonstrated to provide flexible and programmable regulations to control enzymatic DNA circuits and has future applications in DNA computing, biosensing, and DNA storage.
中文翻译:

用于调节 DNA 酶电路的可编程引物切换
开发 DNA 链置换反应 (SDR) 为调节人工核酸电路和网络提供了重要的技术支持。最近,基于酶 SDR 的 DNA 电路因其模块化设计、高正交性信号传导和极快的反应速率而受到广泛关注。典型的酶 SDR 由相对较长的引物(20-30 个核苷酸)调节,这些引物杂交形成稳定的双链结构,促进酶启动事件。由于缺乏方便和简单的引物控制机制,实施更灵活的基于引物的酶促 SDR 法规仍然具有挑战性,从而限制了酶促 DNA 电路的发展。在这项研究中,我们提出了一种称为引物切换调节的方法,通过将可切换电线引入酶电路来实现酶电路的可编程和灵活的调节。我们应用这种方法生成多种酶促 DNA 电路,包括级联、扇入/扇出、双轨、前馈和反馈功能。通过这种方法,只需引入额外的开关线即可实现复杂的电路功能,而无需重构基本电路框架。该方法经实验证明可以提供灵活且可编程的调节来控制酶促 DNA 电路,并在 DNA 计算、生物传感和 DNA 存储方面具有未来的应用。
更新日期:2024-01-29
中文翻译:

用于调节 DNA 酶电路的可编程引物切换
开发 DNA 链置换反应 (SDR) 为调节人工核酸电路和网络提供了重要的技术支持。最近,基于酶 SDR 的 DNA 电路因其模块化设计、高正交性信号传导和极快的反应速率而受到广泛关注。典型的酶 SDR 由相对较长的引物(20-30 个核苷酸)调节,这些引物杂交形成稳定的双链结构,促进酶启动事件。由于缺乏方便和简单的引物控制机制,实施更灵活的基于引物的酶促 SDR 法规仍然具有挑战性,从而限制了酶促 DNA 电路的发展。在这项研究中,我们提出了一种称为引物切换调节的方法,通过将可切换电线引入酶电路来实现酶电路的可编程和灵活的调节。我们应用这种方法生成多种酶促 DNA 电路,包括级联、扇入/扇出、双轨、前馈和反馈功能。通过这种方法,只需引入额外的开关线即可实现复杂的电路功能,而无需重构基本电路框架。该方法经实验证明可以提供灵活且可编程的调节来控制酶促 DNA 电路,并在 DNA 计算、生物传感和 DNA 存储方面具有未来的应用。





















































 京公网安备 11010802027423号
京公网安备 11010802027423号