当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Increasing the Throughput and Reproducibility of Activity-Based Proteome Profiling Studies with Hyperplexing and Intelligent Data Acquisition
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2024-01-22 , DOI: 10.1021/acs.jproteome.3c00598 Hanna G Budayeva 1 , Taylur P Ma 1 , Shuai Wang 2 , Meena Choi 1 , Christopher M Rose 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2024-01-22 , DOI: 10.1021/acs.jproteome.3c00598 Hanna G Budayeva 1 , Taylur P Ma 1 , Shuai Wang 2 , Meena Choi 1 , Christopher M Rose 1
Affiliation
|
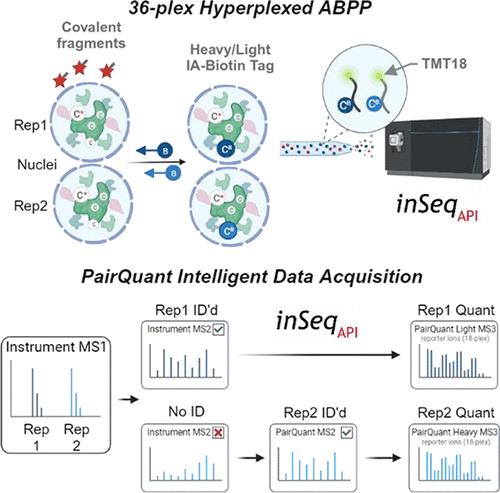
Intelligent data acquisition (IDA) strategies, such as a real-time database search (RTS), have improved the depth of proteome coverage for experiments that utilize isobaric labels and gas phase purification techniques (i.e., SPS-MS3). In this work, we introduce inSeqAPI, an instrument application programing interface (iAPI) program that enables construction of novel data acquisition algorithms. First, we analyze biotinylated cysteine peptides from ABPP experiments to demonstrate that a real-time search method within inSeqAPI performs similarly to an equivalent vendor method. Then, we describe PairQuant, a method within inSeqAPI designed for the hyperplexing approach that utilizes protein-level isotopic labeling and peptide-level TMT labeling. PairQuant allows for TMT analysis of 36 conditions in a single sample and achieves ∼98% coverage of both peptide pair partners in a hyperplexed experiment as well as a 40% improvement in the number of quantified cysteine sites compared with non-RTS acquisition. We applied this method in the ABPP study of ligandable cysteine sites in the nucleus leading to an identification of additional druggable sites on protein- and DNA-interaction domains of transcription regulators and on nuclear ubiquitin ligases.
中文翻译:

通过超复合和智能数据采集提高基于活动的蛋白质组分析研究的通量和重现性
智能数据采集(IDA)策略,例如实时数据库搜索(RTS),提高了利用同量异序标签和气相纯化技术(即SPS-MS3)的实验的蛋白质组覆盖深度。在这项工作中,我们引入了 inSeqAPI,这是一种仪器应用程序编程接口 (iAPI) 程序,可以构建新颖的数据采集算法。首先,我们分析了 ABPP 实验中的生物素化半胱氨酸肽,以证明 inSeqAPI 中的实时搜索方法与等效供应商方法的性能相似。然后,我们描述了 PairQuant,这是 inSeqAPI 中的一种方法,专为利用蛋白质水平同位素标记和肽水平 TMT 标记的超复合方法而设计。 PairQuant 允许对单个样品中的 36 种条件进行 TMT 分析,并在超复合实验中实现两个肽对伙伴的覆盖率约 98%,与非 RTS 采集相比,量化的半胱氨酸位点数量提高了 40%。我们将此方法应用于细胞核中可配体半胱氨酸位点的 ABPP 研究,从而鉴定出转录调节因子的蛋白质和 DNA 相互作用域以及核泛素连接酶上的其他可成药位点。
更新日期:2024-01-22
中文翻译:

通过超复合和智能数据采集提高基于活动的蛋白质组分析研究的通量和重现性
智能数据采集(IDA)策略,例如实时数据库搜索(RTS),提高了利用同量异序标签和气相纯化技术(即SPS-MS3)的实验的蛋白质组覆盖深度。在这项工作中,我们引入了 inSeqAPI,这是一种仪器应用程序编程接口 (iAPI) 程序,可以构建新颖的数据采集算法。首先,我们分析了 ABPP 实验中的生物素化半胱氨酸肽,以证明 inSeqAPI 中的实时搜索方法与等效供应商方法的性能相似。然后,我们描述了 PairQuant,这是 inSeqAPI 中的一种方法,专为利用蛋白质水平同位素标记和肽水平 TMT 标记的超复合方法而设计。 PairQuant 允许对单个样品中的 36 种条件进行 TMT 分析,并在超复合实验中实现两个肽对伙伴的覆盖率约 98%,与非 RTS 采集相比,量化的半胱氨酸位点数量提高了 40%。我们将此方法应用于细胞核中可配体半胱氨酸位点的 ABPP 研究,从而鉴定出转录调节因子的蛋白质和 DNA 相互作用域以及核泛素连接酶上的其他可成药位点。













































 京公网安备 11010802027423号
京公网安备 11010802027423号