当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Special Issue on Artificial Intelligence for Synthetic Biology
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2024-01-12 , DOI: 10.1021/acssynbio.3c00760 Hector García Martín 1, 2, 3, 4 , Stanislav Mazurenko 5, 6 , Huimin Zhao 7
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2024-01-12 , DOI: 10.1021/acssynbio.3c00760 Hector García Martín 1, 2, 3, 4 , Stanislav Mazurenko 5, 6 , Huimin Zhao 7
Affiliation
|
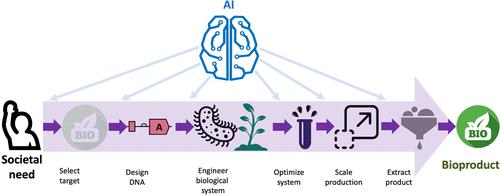
Published as part of the ACS Synthetic Biology virtual special issue “AI for Synthetic Biology”. Synthetic biology presents significant prospects of helping scientists tackle important societal problems. However, a significant hurdle in this endeavor is our inability to predict biological systems as accurately as we predict and simulate physical or chemical ones. This limitation has important fundamental and practical implications: from the practical point of view, we are unable to design biological systems (e.g., proteins, pathways, cells) to a specification (e.g., bind to this molecule with this binding affinity or produce this chemical at this titer, rate, and yield); from the fundamental point of view, we lack an understanding of the underlying mechanisms that produce observed phenotypes. Artificial intelligence (AI) and machine learning (ML) show promise in providing the predictive power that synthetic biology needs and can be applied in all parts of the synthetic biology process (Figure 1). Figure 1. AI can be used to enhance and accelerate synthetic biology in every part of its process: from selecting targets that meet a societal need (e.g., a molecule that binds the histamine H1 receptor involved in allergic responses) to designing the DNA in the pathway needed to produce the desired target, and helping to engineer and optimize biological systems, scale production, and extract the desired product. This Special Issue of ACS Synthetic Biology highlights an extensive range of topics benefiting from AI/ML and a broad spectrum of state-of-the-art AI/ML architectures already being explored in the domain. As this collection demonstrates, a wide variety of biological systems can be targeted using AI/ML. O’Neill et al. (DOI: 10.1021/acssynbio.3c00157) presented a toolkit of signal peptide elements that can be utilized to enhance the production of biopharmaceutical proteins in Chinese hamster ovary cells, leading to ML-assisted vector designs that increase product titers ≥1.8× for a range of products compared to standard industry systems. Khamwachirapithak et al. (DOI: 10.1021/acssynbio.3c00199) applied ML to optimize the bioethanol production in Saccharomyces cerevisiae at ambient and elevated temperatures. After achieving a 63% improvement in ethanol production at 30 °C in the initial round of experiments, an ML-assisted workflow led to an additional 7% improvement at 40 °C. Dallo et al. (DOI: 10.1021/acssynbio.2c00653) studied the design principles for gRNA effectiveness for CRISPRi in the cyanobacterium Synechococcus sp. strain PCC 7002 by constructing, transforming, and analyzing 76 strains using high-density gRNA tiling, correlation analyses, and machine learning. A new software tool DeCOIL was presented by Yang et al. (DOI: 10.1021/acssynbio.3c00301) to optimize degenerate codon libraries for ML-based protein engineering, e.g., to sample promising high-fitness and high-diversity protein variants. The authors demonstrated the utility of the tool on the B1 domain of the immunoglobulin binding protein GB1 and the β-subunit of tryptophan synthase fitness landscapes. Five more studies demonstrated the considerate potential of ML for improving protein engineering pipelines. Marchal et al. (DOI: 10.1021/acssynbio.3c00403) developed an ML-assisted protein engineering workflow based on Gaussian Processes and applied it to improve glycolyl-CoA carboxylases. Among ten variants selected to be tested in vitro, nine were active, marking a dramatic improvement to the prior random mutagenesis success rate. Two novel variants showed a 2-fold increased carboxylation rate and 60% reduced energy demand, respectively. Bricco et al. (DOI: 10.1021/acssynbio.2c00648) leveraged genetic programming to develop the protein engineering tool named POET, demonstrating its utility in engineering novel peptides with improved MRI contrast. The patterns in a high throughput developability data set of ligand scaffold Gp2 protein variants were explored by Golinski et al. (DOI: 10.1021/acssynbio.3c00196) using artificial neural networks, enabling direct visualization of the fitness landscape and revealing evolutionary bottlenecks giving rise to competing subpopulations of sequences with different developability. Chen et al. (DOI: 10.1021/acssynbio.2c00662) performed large-scale sequence–function profiling and epistatic analysis of a flavin mononucleotide-based fluorescent protein CreiLOV, collecting the data for over 90% of single-point and selected combinatorial variants. The authors investigated several statistical and ML models for capturing both specific and global epistasis and concluded that ML-based models were capable of achieving high correlation between the predicted and measured fitness values of higher-order mutants based on the lower-order mutant (1–3 mutations) training data. Kao et al. (DOI: 10.1021/acssynbio.3c00042) employed the deep learning-based model for inverse protein folding, ProteinMPNN to design sequentially divergent ubiquitin variants with high affinities for the HECT domain of E3 ubiquitin-protein ligase Rsp5 exosite, producing several successful designs with increased protein yields, preserved high thermostability, and enhanced binding affinities. Among other exciting applications, Zhang et al. (DOI: 10.1021/acssynbio.3c00225) developed ML models to predict the substrate specificity of three mutants of pyrrolysyl-tRNA synthetase to accept novel noncanonical amino acids. Experimental validation confirmed the activity of 20 out of 24 predicted substrates by at least one PylRS mutant, 11 of which had never been reported to be incorporated into proteins before. The design of universal epitope libraries was studied by Lopez-Martinez et al. (DOI: 10.1021/acssynbio.3c00201), who applied conventional sequence analysis and methods inspired by natural language processing to reveal specific sequence patterns in the linear epitopes deposited in the Immune Epitope Database. Klumpe et al. (DOI: 10.1021/acssynbio.3c00203) demonstrated a considerable potential of deep neural networks for inferring the underlying dynamics of cell response in the presence of measurement noise and stochasticity in the biochemical reactions, using computationally simulated single-cell responses and incorporating different sources of noise and alternative genetic circuit designs. Merzbacher et al. (DOI: 10.1021/acssynbio.3c00120) leveraged Bayesian optimization for the efficient design and optimization of biological circuits. Using several metabolic pathways in E. coli to produce glucaric acid, fatty acids, and p-aminostyrene, the authors illustrated the possible acceleration in screening optimal designs, including the impact of uncertain enzyme kinetic parameters, the use of layered architectures that combine metabolic and genetic control, and multiparameter optimization of a complex model. Several studies of the special issue focus on advanced ML architecture development. Nisonoff et al. (DOI: 10.1021/acssynbio.3c00217) devised a principled probabilistic method to integrate biophysical knowledge into Bayesian neural networks, leading to the models relying more heavily on the biophysical prior information. The authors demonstrated this approach on several examples, including GFP fluorescence and GB1 binding predictions. Van Lent et al. (DOI: 10.1021/acssynbio.3c00186) tackled the combinatorial pathway optimization problem and proposed a mechanistic kinetic model-based framework for the optimization and application of ML methods in iterative metabolic engineering, e.g., for Design–Build–Test–Learn cycles. A new interpretable model architecture Nucleic Transformer, based on self-attention and convolutions, was proposed by He et al. (DOI: 10.1021/acssynbio.3c00154), showcasing its utility in several model tasks, including E. coli promoter classification, viral genome identification, enhancer classification, and chromatin profile predictions. Praljak et al. (DOI: 10.1021/acssynbio.3c00261) proposed ProtWave-VAE, a deep generative model combining the strengths of both multiple sequence alignment-based and autoregressive learning paradigms, to infer meaningful functional and phylogenetic embeddings and tackle downstream protein fitness prediction tasks within alignment-free homologous protein families. Synthetic biology literature was mined and analyzed by two fascinating studies. Xiao et al. (DOI: 10.1021/acssynbio.3c00310) developed a workflow and proposed prompt engineering for the natural language processing tool GPT-4 to extract knowledge from over 170 publications on two oleaginous yeasts. The mined data enabled an ML-based model to predict fermentation titers, showcasing the potential of generative AI in extracting data from research articles to facilitate biomanufacturing development. Meier et al. (DOI: 10.1021/acssynbio.3c00192) used topic modeling to create comprehensive maps of research topics within synthetic biology and coauthorship networks to gain a systematic view of the discipline. Last but not least, the special issue presents a comprehensive review by Yuan et al. (DOI: 10.1021/acssynbio.3c00234), in which the authors discussed the latest advances in developing and applying ML tools for exploring potential natural products, in particular, ML-assisted genome mining of natural products and predicting their bioactivities. The authors also identified several limitations, including the lack of integrated and standardized databases, challenging featurization of natural products, and the lack of robust ML algorithms for small and biased data sets. The special issue thus highlights the extensive use of AI tools in synthetic biology and the considerable promise AI shows in streamlining the workflows and processes in various synthetic biology applications. We believe more and more AI tools will be developed and applied to solve challenging problems in the synthetic biology field in the future. This article has not yet been cited by other publications. Figure 1. AI can be used to enhance and accelerate synthetic biology in every part of its process: from selecting targets that meet a societal need (e.g., a molecule that binds the histamine H1 receptor involved in allergic responses) to designing the DNA in the pathway needed to produce the desired target, and helping to engineer and optimize biological systems, scale production, and extract the desired product.
更新日期:2024-01-12





















































 京公网安备 11010802027423号
京公网安备 11010802027423号