Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Circular DNAzyme-Switched CRISPR/Cas12a Assay for Electrochemiluminescent Response of Demethylase Activity
ACS Sensors ( IF 8.2 ) Pub Date : 2024-01-10 , DOI: 10.1021/acssensors.3c02025 Wei-Wei Yang 1 , Mei-Ling Zhao 1 , Mei-Ling Liu 1 , Wen-Bin Liang 1 , Xia Zhong 1 , Ying Zhuo 1
ACS Sensors ( IF 8.2 ) Pub Date : 2024-01-10 , DOI: 10.1021/acssensors.3c02025 Wei-Wei Yang 1 , Mei-Ling Zhao 1 , Mei-Ling Liu 1 , Wen-Bin Liang 1 , Xia Zhong 1 , Ying Zhuo 1
Affiliation
|
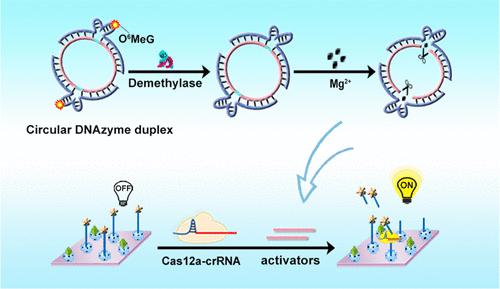
DNA nanostructure provides powerful tools for DNA demethylase activity detection, but its stability has been significantly challenged. By virtue of circular DNA with resistance to exonuclease degradation, herein, the circular DNAzyme duplex with artificial methylated modification was constructed to identify the target and output the DNA activators to drive the CRISPR/Cas12a, constructing an “on–off–on” electrochemiluminescence (ECL) biosensor for monitoring the activity of the O6-methylguanine-DNA methyltransferase (MGMT). Specifically, the circular DNAzyme duplex consisted of the chimeric RNA–DNA substrate ring with double activator sequences and two single-stranded DNAzymes, whose catalytic domains were premodified with the methyl groups. When the MGMT was present, the methylated DNAzymes were repaired and restored the catalytic activity to cleave the chimeric RNA–DNA substrates, followed by the output of DNA activators to initiate the CRISPR/Cas12a. Subsequently, the ECL signals of silver nanoparticle-modified SnO2 nanospheres (Ag@SnO2) were recovered by releasing the ferrocene-labeled quenching probes (Fc-DNA) from the electrode surface because of the trans-cleavage activity of CRISPR/Cas12a, thus achieving the specific and sensitive ECL detection of MGMT from 2.5 × 10–4 to 2.5 × 102 ng/mL with a low limit (9.69 × 10–5 ng/mL). This strategy affords novel ideas and insights into research on how to project stable nucleic acid probes to detect DNA demethylases beyond traditional methods.
中文翻译:

环状 DNAzyme 开关 CRISPR/Cas12a 测定去甲基酶活性的电化学发光反应
DNA纳米结构为DNA去甲基酶活性检测提供了强大的工具,但其稳定性受到了显着挑战。凭借具有抗核酸外切酶降解能力的环状DNA,本文构建了经过人工甲基化修饰的环状DNAzyme双链体,以识别靶标并输出DNA激活剂来驱动CRISPR/Cas12a,构建“开-关-开”电化学发光。 ECL) 生物传感器,用于监测 O 6 -甲基鸟嘌呤-DNA 甲基转移酶 (MGMT) 的活性。具体来说,环状脱氧核糖核酸酶双链体由具有双激活剂序列的嵌合RNA-DNA底物环和两个单链脱氧核糖核酸酶组成,其催化结构域已用甲基进行预修饰。当 MGMT 存在时,甲基化 DNAzyme 被修复并恢复切割嵌合 RNA-DNA 底物的催化活性,随后输出 DNA 激活剂以启动 CRISPR/Cas12a。随后,由于CRISPR/Cas12a的反式切割活性,通过从电极表面释放二茂铁标记的猝灭探针(Fc-DNA)来恢复银纳米颗粒修饰的SnO 2纳米球(Ag@SnO 2 )的ECL信号,从而实现了MGMT从2.5 × 10 –4到2.5 × 10 2 ng/mL的低限(9.69 × 10 –5 ng/mL)的特异性、灵敏的ECL检测。该策略为如何设计稳定的核酸探针来超越传统方法检测 DNA 去甲基酶的研究提供了新颖的想法和见解。
更新日期:2024-01-10
中文翻译:

环状 DNAzyme 开关 CRISPR/Cas12a 测定去甲基酶活性的电化学发光反应
DNA纳米结构为DNA去甲基酶活性检测提供了强大的工具,但其稳定性受到了显着挑战。凭借具有抗核酸外切酶降解能力的环状DNA,本文构建了经过人工甲基化修饰的环状DNAzyme双链体,以识别靶标并输出DNA激活剂来驱动CRISPR/Cas12a,构建“开-关-开”电化学发光。 ECL) 生物传感器,用于监测 O 6 -甲基鸟嘌呤-DNA 甲基转移酶 (MGMT) 的活性。具体来说,环状脱氧核糖核酸酶双链体由具有双激活剂序列的嵌合RNA-DNA底物环和两个单链脱氧核糖核酸酶组成,其催化结构域已用甲基进行预修饰。当 MGMT 存在时,甲基化 DNAzyme 被修复并恢复切割嵌合 RNA-DNA 底物的催化活性,随后输出 DNA 激活剂以启动 CRISPR/Cas12a。随后,由于CRISPR/Cas12a的反式切割活性,通过从电极表面释放二茂铁标记的猝灭探针(Fc-DNA)来恢复银纳米颗粒修饰的SnO 2纳米球(Ag@SnO 2 )的ECL信号,从而实现了MGMT从2.5 × 10 –4到2.5 × 10 2 ng/mL的低限(9.69 × 10 –5 ng/mL)的特异性、灵敏的ECL检测。该策略为如何设计稳定的核酸探针来超越传统方法检测 DNA 去甲基酶的研究提供了新颖的想法和见解。






























 京公网安备 11010802027423号
京公网安备 11010802027423号