当前位置:
X-MOL 学术
›
Comput. Biol. Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Application of ensemble learning for predicting GABAA receptor agonists
Computers in Biology and Medicine ( IF 7.0 ) Pub Date : 2024-01-03 , DOI: 10.1016/j.compbiomed.2024.107958 Fu Xiao 1 , Xiaoyu Ding 2 , Yan Shi 3 , Dingyan Wang 2 , Yitian Wang 2 , Chen Cui 2 , Tingfei Zhu 2 , Kaixian Chen 4 , Ping Xiang 3 , Xiaomin Luo 4
Computers in Biology and Medicine ( IF 7.0 ) Pub Date : 2024-01-03 , DOI: 10.1016/j.compbiomed.2024.107958 Fu Xiao 1 , Xiaoyu Ding 2 , Yan Shi 3 , Dingyan Wang 2 , Yitian Wang 2 , Chen Cui 2 , Tingfei Zhu 2 , Kaixian Chen 4 , Ping Xiang 3 , Xiaomin Luo 4
Affiliation
|
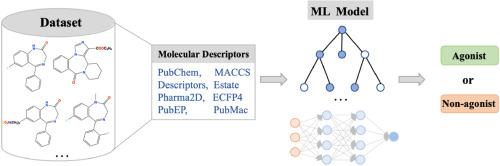
Over the past few decades, agonists binding to the benzodiazepine site of the GABA receptor have been successfully developed as clinical drugs. Different modulators (agonist, antagonist, and reverse agonist) bound to benzodiazepine sites exhibit different or even opposite pharmacological effects, however, their structures are so similar that it is difficult to distinguish them based solely on molecular skeleton. This study aims to develop classification models for predicting the agonists. 306 agonists or non-agonists were collected from literature. Six machine learning algorithms including RF, XGBoost, AdaBoost, GBoost, SVM, and ANN algorithms were employed for model development. Using six descriptors including 1D/2D Descriptors, ECFP4, 2D-Pharmacophore, MACCS, PubChem, and Estate fingerprint to characterize chemical structures. The model interpretability was explored by SHAP method. The best model demonstrated an AUC value of 0.905 and an MCC value of 0.808 for the test set. The PubMac-based model (PubMac-GB) achieved best AUC values of 0.935 for test set. The SHAP analysis results emphasized that MaccsFP62, ECFP_624, ECFP_724, and PubchemFP213 were the crucial molecular features. Applicability domain analysis was also performed to determine reliable prediction boundaries for the model. The PubMac-GB model was applied to virtual screening for potential GABA agonists and the top 100 compounds were given. Overall, our ensemble learning-based model (PubMac-GB) achieved comparable performance and would be helpful in effectively identifying agonists of GABA receptors.
中文翻译:

集成学习在预测 GABAA 受体激动剂中的应用
在过去的几十年里,与 GABA 受体的苯二氮卓类药物位点结合的激动剂已成功开发为临床药物。与苯二氮卓类药物位点结合的不同调节剂 (激动剂、拮抗剂和反向激动剂) 表现出不同甚至相反的药理作用,但是,它们的结构非常相似,以至于很难仅根据分子骨架来区分它们。本研究旨在开发用于预测激动剂的分类模型。从文献中收集了 306 种激动剂或非激动剂。模型开发采用了 6 种机器学习算法,包括 RF、XGBoost、AdaBoost、GBoost、SVM 和 ANN 算法。使用六种描述符(包括 1D/2D 描述符、ECFP4、2D-药效团、MACCS、PubChem 和 Estate 指纹图谱)来表征化学结构。通过 SHAP 方法探索模型的可解释性。最佳模型显示测试集的 AUC 值为 0.905,MCC 值为 0.808。基于 PubMac 的模型 (PubMac-GB) 在测试集上实现了 0.935 的最佳 AUC 值。SHAP 分析结果强调 MaccsFP62 、 ECFP_624 、 ECFP_724 和 PubchemFP213 是关键的分子特征。还进行了适用性域分析以确定模型的可靠预测边界。将 PubMac-GB 模型应用于潜在 GABA 激动剂的虚拟筛选,并给出前 100 种化合物。总体而言,我们基于集成学习的模型 (PubMac-GB) 取得了相当的性能,将有助于有效识别 GABA 受体的激动剂。
更新日期:2024-01-03
中文翻译:

集成学习在预测 GABAA 受体激动剂中的应用
在过去的几十年里,与 GABA 受体的苯二氮卓类药物位点结合的激动剂已成功开发为临床药物。与苯二氮卓类药物位点结合的不同调节剂 (激动剂、拮抗剂和反向激动剂) 表现出不同甚至相反的药理作用,但是,它们的结构非常相似,以至于很难仅根据分子骨架来区分它们。本研究旨在开发用于预测激动剂的分类模型。从文献中收集了 306 种激动剂或非激动剂。模型开发采用了 6 种机器学习算法,包括 RF、XGBoost、AdaBoost、GBoost、SVM 和 ANN 算法。使用六种描述符(包括 1D/2D 描述符、ECFP4、2D-药效团、MACCS、PubChem 和 Estate 指纹图谱)来表征化学结构。通过 SHAP 方法探索模型的可解释性。最佳模型显示测试集的 AUC 值为 0.905,MCC 值为 0.808。基于 PubMac 的模型 (PubMac-GB) 在测试集上实现了 0.935 的最佳 AUC 值。SHAP 分析结果强调 MaccsFP62 、 ECFP_624 、 ECFP_724 和 PubchemFP213 是关键的分子特征。还进行了适用性域分析以确定模型的可靠预测边界。将 PubMac-GB 模型应用于潜在 GABA 激动剂的虚拟筛选,并给出前 100 种化合物。总体而言,我们基于集成学习的模型 (PubMac-GB) 取得了相当的性能,将有助于有效识别 GABA 受体的激动剂。































 京公网安备 11010802027423号
京公网安备 11010802027423号