当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Ultrahigh-Throughput Directed Evolution of Polymer-Degrading Enzymes Using Yeast Display
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2023-12-05 , DOI: 10.1021/jacs.3c08291 Mario A Cribari 1 , Maxwell J Unger 1 , Ilona C Unarta 1, 2 , Ashley N Ogorek 1 , Xuhui Huang 1, 2 , Jeffrey D Martell 1, 3
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2023-12-05 , DOI: 10.1021/jacs.3c08291 Mario A Cribari 1 , Maxwell J Unger 1 , Ilona C Unarta 1, 2 , Ashley N Ogorek 1 , Xuhui Huang 1, 2 , Jeffrey D Martell 1, 3
Affiliation
|
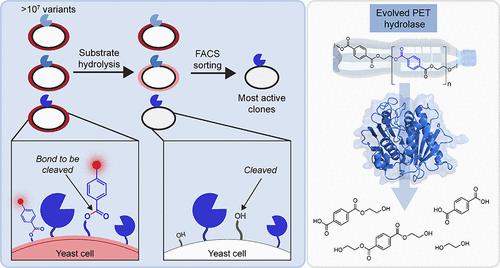
Enzymes that degrade synthetic polymers have attracted intense interest for eco-friendly plastic recycling. However, because enzymes did not evolve for the cleavage of abiotic polymers, directed evolution strategies are needed to enhance activity for plastic degradation. Previous directed evolution efforts relied on polymer degradation assays that were limited to screening ∼104 mutants. Here, we report a high-throughput yeast surface display platform to rapidly evaluate >107 enzyme mutants for increased activity in cleaving synthetic polymers. In this platform, individual yeast cells display distinct mutants, and enzyme activity is detected by a change in fluorescence upon the cleavage of a synthetic probe resembling a polymer of interest. Highly active mutants are isolated by fluorescence activated cell sorting and identified through DNA sequencing. To demonstrate this platform, we performed directed evolution of a polyethylene terephthalate (PET)-depolymerizing enzyme, leaf and branch compost cutinase (LCC). We identified activity-boosting mutations that substantially increased the kinetics of degradation of solid PET films. Biochemical assays and molecular dynamics (MD) simulations of the most active variants suggest that the H218Y mutation improves the binding of the enzyme to PET. Overall, this evolution platform increases the screening throughput of polymer-degrading enzymes by 3 orders of magnitude and identifies mutations that enhance kinetics for depolymerizing solid substrates.
中文翻译:

使用酵母展示进行聚合物降解酶的超高通量定向进化
降解合成聚合物的酶引起了人们对环保塑料回收的浓厚兴趣。然而,由于酶并不是为了裂解非生物聚合物而进化的,因此需要定向进化策略来增强塑料降解的活性。先前的定向进化工作依赖于聚合物降解测定,该测定仅限于筛选~10 4突变体。在这里,我们报告了一个高通量酵母表面展示平台,可快速评估> 10 7 个酶突变体,以提高裂解合成聚合物的活性。在该平台中,单个酵母细胞表现出不同的突变体,并且通过类似目标聚合物的合成探针裂解时荧光的变化来检测酶活性。通过荧光激活细胞分选分离高活性突变体,并通过 DNA 测序进行鉴定。为了演示这个平台,我们对聚对苯二甲酸乙二醇酯(PET)解聚酶、叶和树枝堆肥角质酶(LCC)进行了定向进化。我们发现了活性增强突变,大大增加了固体 PET 薄膜的降解动力学。最活跃变体的生化测定和分子动力学 (MD) 模拟表明 H218Y 突变提高了酶与 PET 的结合。总体而言,该进化平台将聚合物降解酶的筛选通量提高了 3 个数量级,并识别出增强固体底物解聚动力学的突变。
更新日期:2023-12-05
中文翻译:

使用酵母展示进行聚合物降解酶的超高通量定向进化
降解合成聚合物的酶引起了人们对环保塑料回收的浓厚兴趣。然而,由于酶并不是为了裂解非生物聚合物而进化的,因此需要定向进化策略来增强塑料降解的活性。先前的定向进化工作依赖于聚合物降解测定,该测定仅限于筛选~10 4突变体。在这里,我们报告了一个高通量酵母表面展示平台,可快速评估> 10 7 个酶突变体,以提高裂解合成聚合物的活性。在该平台中,单个酵母细胞表现出不同的突变体,并且通过类似目标聚合物的合成探针裂解时荧光的变化来检测酶活性。通过荧光激活细胞分选分离高活性突变体,并通过 DNA 测序进行鉴定。为了演示这个平台,我们对聚对苯二甲酸乙二醇酯(PET)解聚酶、叶和树枝堆肥角质酶(LCC)进行了定向进化。我们发现了活性增强突变,大大增加了固体 PET 薄膜的降解动力学。最活跃变体的生化测定和分子动力学 (MD) 模拟表明 H218Y 突变提高了酶与 PET 的结合。总体而言,该进化平台将聚合物降解酶的筛选通量提高了 3 个数量级,并识别出增强固体底物解聚动力学的突变。






























 京公网安备 11010802027423号
京公网安备 11010802027423号