当前位置:
X-MOL 学术
›
Environ. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
DNA metabarcoding focused on difficult-to-culture protists: An effective approach to clarify biological interactions
Environmental Microbiology ( IF 4.3 ) Pub Date : 2023-10-18 , DOI: 10.1111/1462-2920.16524
Yasuhide Nakamura 1, 2 , Hiryori Itagaki 3 , Akihiro Tuji 2 , Shinji Shimode 4 , Atsushi Yamaguchi 5 , Kiyotaka Hidaka 6 , Eri Ogiso-Tanaka 7
Environmental Microbiology ( IF 4.3 ) Pub Date : 2023-10-18 , DOI: 10.1111/1462-2920.16524
Yasuhide Nakamura 1, 2 , Hiryori Itagaki 3 , Akihiro Tuji 2 , Shinji Shimode 4 , Atsushi Yamaguchi 5 , Kiyotaka Hidaka 6 , Eri Ogiso-Tanaka 7
Affiliation
|
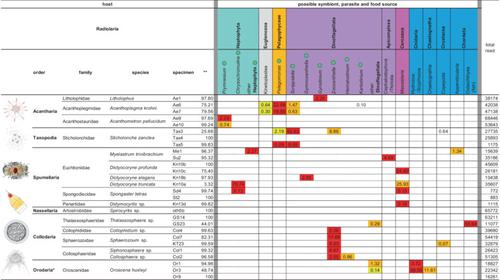
DNA metabarcoding on a single organism is a promising approach to clarify the biological interactions (e.g., predator–prey relationships and symbiosis, including parasitism) of difficult-to-culture protists. To evaluate the effectiveness of this method, Radiolaria and Phaeodaria, which are ecologically important protistan groups, were chosen as target taxa. DNA metabarcoding on a single organism focused on the V9 region of the 18S rRNA gene revealed potential symbionts, parasites and food sources of Radiolaria and Phaeodaria. Previously reported hosts and symbionts (parasites) were detected, and newly recognized combinations were also identified. The contained organisms largely differed between Radiolaria and Phaeodaria. In Radiolaria, members of the same order tended to contain similar organisms, and the taxonomic composition of possible symbionts, parasites, and food sources was fixed at the species level. Members of the same phaeodarian family, however, did not contain similar organisms, and body part (i.e., the central capsule or the phaeodium) was the most important factor that divided the taxonomic composition of detected organisms, implying that the selection of appropriate body part is important when trying to ascertain contained organisms, even for unicellular zooplankton. Our results show that DNA metabarcoding on a single organism is effective in revealing the biological interactions of difficult-to-culture protists.
中文翻译:

DNA 元条形码聚焦于难以培养的原生生物:阐明生物相互作用的有效方法
单个生物体上的 DNA 元条形码是阐明难以培养的原生生物的生物相互作用(例如捕食者-猎物关系和共生,包括寄生)的一种有前景的方法。为了评估该方法的有效性,选择生态上重要的原生生物类群放射虫和褐藻作为目标类群。对单一生物体 18S rRNA 基因 V9 区域的 DNA 元条形码揭示了放射虫和褐虫的潜在共生体、寄生虫和食物来源。检测到了先前报道的宿主和共生体(寄生虫),并且还鉴定了新识别的组合。放射虫和褐虫所包含的生物体差异很大。在放射虫中,同一目的成员往往包含相似的生物体,并且可能的共生体、寄生虫和食物来源的分类组成在物种水平上是固定的。然而,同一Phaeodarian科的成员并不包含相似的生物体,并且身体部位(即中央胶囊或Phaeodium)是划分检测到的生物分类组成的最重要因素,这意味着选择适当的身体部位当试图确定所包含的生物体时,即使是单细胞浮游动物,这一点也很重要。我们的结果表明,单个生物体上的 DNA 元条形码可有效揭示难以培养的原生生物的生物相互作用。
更新日期:2023-10-18
中文翻译:

DNA 元条形码聚焦于难以培养的原生生物:阐明生物相互作用的有效方法
单个生物体上的 DNA 元条形码是阐明难以培养的原生生物的生物相互作用(例如捕食者-猎物关系和共生,包括寄生)的一种有前景的方法。为了评估该方法的有效性,选择生态上重要的原生生物类群放射虫和褐藻作为目标类群。对单一生物体 18S rRNA 基因 V9 区域的 DNA 元条形码揭示了放射虫和褐虫的潜在共生体、寄生虫和食物来源。检测到了先前报道的宿主和共生体(寄生虫),并且还鉴定了新识别的组合。放射虫和褐虫所包含的生物体差异很大。在放射虫中,同一目的成员往往包含相似的生物体,并且可能的共生体、寄生虫和食物来源的分类组成在物种水平上是固定的。然而,同一Phaeodarian科的成员并不包含相似的生物体,并且身体部位(即中央胶囊或Phaeodium)是划分检测到的生物分类组成的最重要因素,这意味着选择适当的身体部位当试图确定所包含的生物体时,即使是单细胞浮游动物,这一点也很重要。我们的结果表明,单个生物体上的 DNA 元条形码可有效揭示难以培养的原生生物的生物相互作用。

































 京公网安备 11010802027423号
京公网安备 11010802027423号