Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
OSMAC Approach and Cocultivation for the Induction of Secondary Metabolism of the Fungus Pleotrichocladium opacum
ACS Omega ( IF 3.7 ) Pub Date : 2023-10-16 , DOI: 10.1021/acsomega.3c06299 Víctor Rodríguez Martín-Aragón 1 , Mónica Trigal Martínez 1 , Cristina Cuadrado 2 , Antonio Hernández Daranas 2 , Antonio Fernández Medarde 1 , José M Sánchez López 1
ACS Omega ( IF 3.7 ) Pub Date : 2023-10-16 , DOI: 10.1021/acsomega.3c06299 Víctor Rodríguez Martín-Aragón 1 , Mónica Trigal Martínez 1 , Cristina Cuadrado 2 , Antonio Hernández Daranas 2 , Antonio Fernández Medarde 1 , José M Sánchez López 1
Affiliation
|
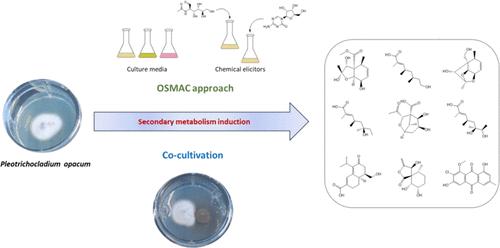
The application of an OSMAC (One Strain–Many Compounds) approach on the fungus Pleotrichocladium opacum, isolated from a soil sample collected on the coast of Asturias (Spain), using different culture media, chemical elicitors, and cocultivation techniques resulted in the isolation and identification of nine new compounds (8, 9, 12, 15–18, 20, 21), along with 15 known ones (1–7, 10, 11, 14, 19, 22–25). Compounds 1–9 were detected in fungal extracts from JSA liquid fermentation, compounds 10–12 were isolated from a solid rice medium, whereas compounds 14 and 15 were isolated from a solid wheat medium. Addition of 5-azacytidine to the solid rice medium caused the accumulation of compounds 16–18, whereas adding N-acetyl-d-glucosamine triggered the production of two additional metabolites, 19 and 20. Finally, cocultivation of the fungus Pleotrichocladium opacum with Echinocatena sp. in a solid PDA medium led to the production of five additional natural products, 21–25. The structures of the new compounds were elucidated by HRESIMS and 1D and 2D NMR as well as by comparison with literature data. DP4+ and mix-J-DP4 computational methods were applied to determine the relative configurations of the novel compounds, and in some cases, the absolute configurations were assigned by a comparison of the optical rotations with those of related natural products.
中文翻译:

OSMAC 方法和共培养诱导真菌 Pleotrichocladium opacum 的次生代谢
对从阿斯图里亚斯(西班牙)海岸收集的土壤样本中分离出的真菌 Pleotrichocladium opacum 应用OSMAC(一株多种化合物)方法,使用不同的培养基、化学引发剂和共培养技术,得到了分离和鉴定了9种新化合物( 8、9、12、15 – 18、20、21 ),以及15种已知化合物( 1 – 7、10、11、14、19、22 – 25 ) 。_ _ 在 JSA 液体发酵的真菌提取物中检测到化合物 1 – 9,从固体稻米培养基中分离出化合物10 – 12 ,而从固体小麦培养基中分离出化合物14和15 。向固体稻米培养基中添加 5-氮杂胞苷导致化合物16 – 18的积累,而添加N-乙酰基-d-葡萄糖胺则引发两种额外代谢物19和20的产生。最后,真菌Pleotrichocladium opacum与Echinocatena sp 的共培养。在固体 PDA 培养基中导致另外五种天然产物的产生,21 – 25。通过 HRESIMS、1D 和 2D NMR 以及与文献数据的比较,阐明了新化合物的结构。应用DP4+和mix - J -DP4计算方法来确定新化合物的相对构型,在某些情况下,通过与相关天然产物的旋光度比较来确定绝对构型。
更新日期:2023-10-16
中文翻译:

OSMAC 方法和共培养诱导真菌 Pleotrichocladium opacum 的次生代谢
对从阿斯图里亚斯(西班牙)海岸收集的土壤样本中分离出的真菌 Pleotrichocladium opacum 应用OSMAC(一株多种化合物)方法,使用不同的培养基、化学引发剂和共培养技术,得到了分离和鉴定了9种新化合物( 8、9、12、15 – 18、20、21 ),以及15种已知化合物( 1 – 7、10、11、14、19、22 – 25 ) 。_ _ 在 JSA 液体发酵的真菌提取物中检测到化合物 1 – 9,从固体稻米培养基中分离出化合物10 – 12 ,而从固体小麦培养基中分离出化合物14和15 。向固体稻米培养基中添加 5-氮杂胞苷导致化合物16 – 18的积累,而添加N-乙酰基-d-葡萄糖胺则引发两种额外代谢物19和20的产生。最后,真菌Pleotrichocladium opacum与Echinocatena sp 的共培养。在固体 PDA 培养基中导致另外五种天然产物的产生,21 – 25。通过 HRESIMS、1D 和 2D NMR 以及与文献数据的比较,阐明了新化合物的结构。应用DP4+和mix - J -DP4计算方法来确定新化合物的相对构型,在某些情况下,通过与相关天然产物的旋光度比较来确定绝对构型。






































 京公网安备 11010802027423号
京公网安备 11010802027423号