当前位置:
X-MOL 学术
›
Bioconjugate Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Solid-Phase Collateral Cleavage System Based on CRISPR/Cas12 and Its Application toward Facile One-Pot Multiplex Double-Stranded DNA Detection
Bioconjugate Chemistry ( IF 4.0 ) Pub Date : 2023-10-02 , DOI: 10.1021/acs.bioconjchem.3c00294
Hiroki Shigemori 1, 2 , Satoshi Fujita 1 , Eiichi Tamiya 1, 3 , Shin-Ichi Wakida 1, 3 , Hidenori Nagai 1, 2
Bioconjugate Chemistry ( IF 4.0 ) Pub Date : 2023-10-02 , DOI: 10.1021/acs.bioconjchem.3c00294
Hiroki Shigemori 1, 2 , Satoshi Fujita 1 , Eiichi Tamiya 1, 3 , Shin-Ichi Wakida 1, 3 , Hidenori Nagai 1, 2
Affiliation
|
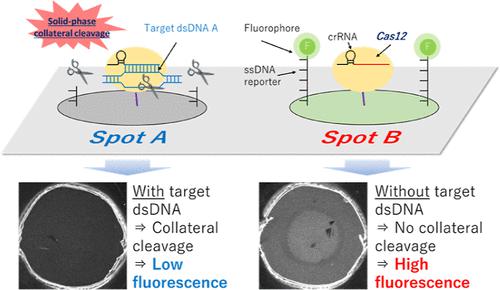
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 12 (Cas12) system is attracting interest for its potential as a next-generation nucleic acid detection tool. The system can recognize double-stranded DNA (dsDNA) based on Cas12-CRISPR RNA (crRNA) and induce signal transduction by collateral cleavage. This property is expected to simplify comprehensive genotyping. Here, we report a solid-phase collateral cleavage (SPCC) reaction by CRISPR/Cas12 and its application toward one-pot multiplex dsDNA detection with minimal operational steps. In the sensor, Cas12-crRNA and single-stranded DNA (ssDNA) are immobilized on the sensing surface and act as enzyme and reporter substrates, respectively. We also report a dual-target dsDNA sensor prepared by immobilizing Cas12-crRNA and a fluorophore-labeled ssDNA reporter on separate spots. When a spot captures a target dsDNA sequence, it cleaves the ssDNA reporter on the same spot and reduces its fluorescence by 42.1–57.3%. Crucially, spots targeting different sequences do not show a reduction in fluorescence, thus confirming the one-pot multiplex dsDNA detection by SPCC. Furthermore, the sequence specificity has a two-base resolution, and the detectable concentration for the target dsDNA is at least 10–9 M. In the future, the SPCC-based sensor array could achieve one-pot comprehensive genotyping by using an array spotter as a reagent-immobilizing method.
中文翻译:

基于 CRISPR/Cas12 的固相侧切系统及其在一锅法多重双链 DNA 检测中的应用
成簇规则间隔短回文重复序列 (CRISPR)/CRISPR 相关蛋白 12 (Cas12) 系统因其作为下一代核酸检测工具的潜力而引起人们的兴趣。该系统可以识别基于Cas12-CRISPR RNA (crRNA)的双链DNA (dsDNA),并通过旁路切割诱导信号转导。该特性有望简化综合基因分型。在这里,我们报告了 CRISPR/Cas12 的固相侧向裂解 (SPCC) 反应及其在以最少操作步骤进行一锅多重 dsDNA 检测中的应用。在传感器中,Cas12-crRNA 和单链 DNA (ssDNA) 固定在传感表面上,分别充当酶和报告底物。我们还报道了通过将 Cas12-crRNA 和荧光团标记的 ssDNA 报告基因固定在不同的点上而制备的双靶点 dsDNA 传感器。当一个点捕获目标 dsDNA 序列时,它会切割同一点上的 ssDNA 报告基因,并将其荧光降低 42.1–57.3%。至关重要的是,针对不同序列的斑点并未表现出荧光减少,从而证实了 SPCC 的一锅多重 dsDNA 检测。此外,序列特异性具有两个碱基的分辨率,目标双链DNA的可检测浓度至少为10 – 9 M。未来,基于SPCC的传感器阵列可以通过阵列点样器实现一锅综合基因分型作为试剂固定化方法。
更新日期:2023-10-02
中文翻译:

基于 CRISPR/Cas12 的固相侧切系统及其在一锅法多重双链 DNA 检测中的应用
成簇规则间隔短回文重复序列 (CRISPR)/CRISPR 相关蛋白 12 (Cas12) 系统因其作为下一代核酸检测工具的潜力而引起人们的兴趣。该系统可以识别基于Cas12-CRISPR RNA (crRNA)的双链DNA (dsDNA),并通过旁路切割诱导信号转导。该特性有望简化综合基因分型。在这里,我们报告了 CRISPR/Cas12 的固相侧向裂解 (SPCC) 反应及其在以最少操作步骤进行一锅多重 dsDNA 检测中的应用。在传感器中,Cas12-crRNA 和单链 DNA (ssDNA) 固定在传感表面上,分别充当酶和报告底物。我们还报道了通过将 Cas12-crRNA 和荧光团标记的 ssDNA 报告基因固定在不同的点上而制备的双靶点 dsDNA 传感器。当一个点捕获目标 dsDNA 序列时,它会切割同一点上的 ssDNA 报告基因,并将其荧光降低 42.1–57.3%。至关重要的是,针对不同序列的斑点并未表现出荧光减少,从而证实了 SPCC 的一锅多重 dsDNA 检测。此外,序列特异性具有两个碱基的分辨率,目标双链DNA的可检测浓度至少为10 – 9 M。未来,基于SPCC的传感器阵列可以通过阵列点样器实现一锅综合基因分型作为试剂固定化方法。

































 京公网安备 11010802027423号
京公网安备 11010802027423号