Computational Biology and Chemistry Pub Date : 2023-09-26 , DOI: 10.1016/j.compbiolchem.2023.107962 Helmi Husaini Zainal Fithri 1 , Zalikha Ibrahim 2 , Ernie Zuraida Ali 3
|
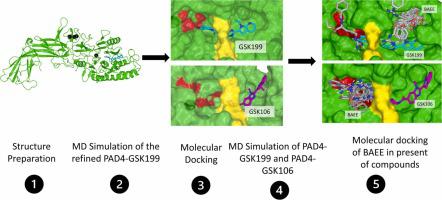
Protein arginine deiminase IV (PAD4) is a potential target for diseases including rheumatoid arthritis and cancers. Currently, GSK199 is a potent, selective yet reversible PAD4 inhibitor. Its derivative, GSK106, on the other hand, was reported as an inactive compound when tested against PAD4 assay. Although they had similar skeleton, their impact towards PAD4 structural and flexibility is unknown. In order to fill the research gap, the impact of GSK199 and GSK106 binding towards PAD4 stability and flexibility is investigated via a combination of computational methods. Molecular docking indicates that GSK199 and GSK106 are capable to bind at PAD4 pocket by using its back door with −10.6 kcal/mol and −9.6 kcal/mol, respectively. The simulations of both complexes were stable throughout 100 ns. The structure of PAD4 exhibited a tighter packing in the presence of GSK106 compared to GSK199. The RMSF analysis demonstrates significant changes between the PAD4-GSK199 and PAD4-GSK106 simulations in the regions containing residues 136, 160, 220, 438, and 606. The Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) analysis shows a marked difference in binding free energies, with −11.339 kcal/mol for the PAD4-GSK199 complex and 1.063 kcal/mol for the PAD4-GSK106 complex. The hydrogen bond analysis revealed that the GSK199 and GSK106 binding to PAD4 are assisted by six hydrogen bonds and three hydrogen bonds, respectively. The visualisation of the MD simulations revealed that GSK199 remained in the PAD4 pocket, whereas GSK106 shifted away from the catalytic site. Meanwhile, molecular dockings of benzoyl arginine amide (BAEE) substrate have shown that BAEE is able to bind to PAD4 catalytic site when GSK106 was present but not when GSK199 occupied the site. Overall, combination of computational approaches successfully described the behaviour of binding pocket of PAD4 structure in the presence of the active and inactive compounds.
中文翻译:

GSK199 和 GSK106 结合对蛋白质精氨酸脱亚胺酶 IV 稳定性和灵活性的影响:一种计算方法
蛋白质精氨酸脱亚胺酶IV (PAD4) 是类风湿关节炎和癌症等疾病的潜在靶标。目前,GSK199 是一种有效、选择性且可逆的 PAD4 抑制剂。另一方面,其衍生物 GSK106 在 PAD4 检测中被报告为无活性化合物。尽管它们具有相似的骨架,但它们对 PAD4 结构和灵活性的影响尚不清楚。为了填补研究空白,通过组合计算方法研究了 GSK199 和 GSK106 结合对 PAD4 稳定性和灵活性的影响。分子对接表明GSK199和GSK106能够通过其后门分别以-10.6 kcal/mol和-9.6 kcal/mol与PAD4口袋结合。两种复合物的模拟在 100 ns 内保持稳定。与 GSK199 相比,PAD4 的结构在 GSK106 存在下表现出更紧密的堆积。RMSF 分析表明,PAD4-GSK199 和 PAD4-GSK106 模拟在包含残基 136、160、220、438 和 606 的区域中存在显着变化。分子力学泊松-玻尔兹曼表面积 (MMPBSA) 分析显示结合存在显着差异自由能,PAD4-GSK199 复合物的自由能为-11.339 kcal/mol,PAD4-GSK106 复合物的自由能为1.063 kcal/mol。氢键分析表明GSK199和GSK106与PAD4的结合分别受到六个氢键和三个氢键的辅助。MD 模拟的可视化显示 GSK199 保留在 PAD4 口袋中,而 GSK106 则移离催化位点。同时,苯甲酰精氨酸酰胺(BAEE)底物的分子对接表明,当GSK106存在时,BAEE能够与PAD4催化位点结合,但当GSK199占据该位点时则不能结合。总体而言,计算方法的组合成功地描述了在活性和非活性化合物存在的情况下 PAD4 结构的结合口袋的行为。































 京公网安备 11010802027423号
京公网安备 11010802027423号