Water Research ( IF 11.4 ) Pub Date : 2023-09-29 , DOI: 10.1016/j.watres.2023.120682 Chen Wang 1 , Huiying Yang 1 , Huafeng Liu 1 , Xu-Xiang Zhang 2 , Liping Ma 1
|
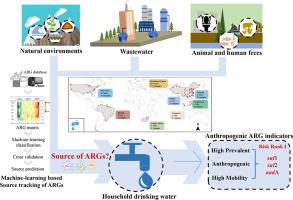
Although the presence of antibiotic resistance genes (ARGs) in drinking water and their potential horizontal gene transfer to pathogenic microbes are known to pose a threat to human health, their pollution levels and potential anthropogenic sources are poorly understood. In this study, broad-spectrum ARG profiling combined with machine-learning-based source classification SourceTracker was performed to investigate the pollution sources of ARGs in household drinking water collected from 95 households in 47 cities of eight countries/regions. In total, 451 ARG subtypes belonging to 19 ARG types were detected with total abundance in individual samples ranging from 1.4 × 10−4 to 1.5 × 10° copies per cell. Source tracking analysis revealed that many ARGs were highly contributed by anthropogenic sources (37.1%), mainly wastewater treatment plants. The regions with the highest detected ARG contribution from wastewater (∼84.3%) used recycled water as drinking water, indicating the need for better ARG control strategies to ensure safe water quality in these regions. Among ARG types, sulfonamide, rifamycin and tetracycline resistance genes were mostly anthropogenic in origin. The contributions of anthropogenic sources to the 20 core ARGs detected in all of the studied countries/regions varied from 36.6% to 84.1%. Moreover, the anthropogenic contribution of 17 potential mobile ARGs identified in drinking water was significantly higher than other ARGs, and metagenomic assembly revealed that these mobile ARGs were carried by diverse potential pathogens. These results indicate that human activities have exacerbated the constant input and transmission of ARGs in drinking water. Our further risk classification framework revealed three ARGs (sul1, sul2 and aadA) that pose the highest risk to public health given their high prevalence, anthropogenic sources and mobility, facilitating accurate monitoring and control of anthropogenic pollution in drinking water.
中文翻译:

基于机器学习的源头追踪揭示了人为对家庭饮用水抗生素抗性基因污染的贡献
尽管已知饮用水中抗生素抗性基因(ARG)的存在及其向病原微生物的潜在水平基因转移对人类健康构成威胁,但对其污染水平和潜在的人为来源知之甚少。本研究采用广谱ARG分析与基于机器学习的来源分类SourceTracker相结合,对8个国家/地区47个城市的95户家庭饮用水中ARG的污染源进行了调查。总共检测到属于 19 个 ARG 类型的 451 个 ARG 亚型,各个样本中的总丰度范围为每个细胞 1.4 × 10 -4至 1.5 × 10° 拷贝。来源追踪分析显示,许多ARGs很大程度上来自人为来源(37.1%),主要是废水处理厂。废水中检测到的 ARG 贡献最高的地区 (∼84.3%) 使用再生水作为饮用水,表明需要更好的 ARG 控制策略来确保这些地区的水质安全。在ARG类型中,磺酰胺、利福霉素和四环素抗性基因大多是人为起源的。在所有研究国家/地区检测到的 20 种核心 ARG 中,人为来源的贡献率从 36.6% 到 84.1% 不等。此外,饮用水中鉴定出的 17 种潜在移动 ARG 的人为贡献显着高于其他 ARG,宏基因组组装表明这些移动 ARG 由多种潜在病原体携带。这些结果表明,人类活动加剧了饮用水中ARG的持续输入和传播。我们进一步的风险分类框架揭示了三种 ARG(sul 1、sul 2 和aad A),由于其高患病率、人为来源和流动性,对公共卫生构成最高风险,有助于准确监测和控制饮用水中的人为污染。































 京公网安备 11010802027423号
京公网安备 11010802027423号