当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
High-Throughput Deconvolution of Native Protein Mass Spectrometry Imaging Data Sets for Mass Domain Analysis
Analytical Chemistry ( IF 6.7 ) Pub Date : 2023-09-06 , DOI: 10.1021/acs.analchem.3c02616 Oliver J Hale 1 , Helen J Cooper 1 , Michael T Marty 2
Analytical Chemistry ( IF 6.7 ) Pub Date : 2023-09-06 , DOI: 10.1021/acs.analchem.3c02616 Oliver J Hale 1 , Helen J Cooper 1 , Michael T Marty 2
Affiliation
|
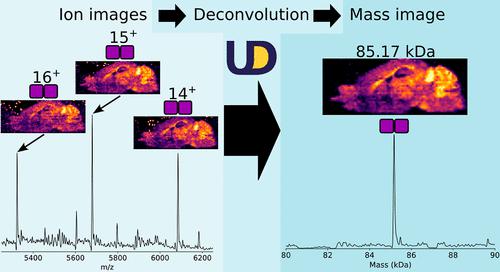
Protein mass spectrometry imaging (MSI) with electrospray-based ambient ionization techniques, such as nanospray desorption electrospray ionization (nano-DESI), generates data sets in which each pixel corresponds to a mass spectrum populated by peaks corresponding to multiply charged protein ions. Importantly, the signal associated with each protein is split among multiple charge states. These peaks can be transformed into the mass domain by spectral deconvolution. When proteins are imaged under native/non-denaturing conditions to retain non-covalent interactions, deconvolution is particularly valuable in helping interpret the data. To improve the acquisition speed, signal-to-noise ratio, and sensitivity, native MSI is usually performed using mass resolving powers that do not provide isotopic resolution, and conventional algorithms for deconvolution of lower-resolution data are not suitable for these large data sets. UniDec was originally developed to enable rapid deconvolution of complex protein mass spectra. Here, we developed an updated feature set harnessing the high-throughput module, MetaUniDec, to deconvolve each pixel of native MSI data sets and transform m/z-domain image files to the mass domain. New tools enable the reading, processing, and output of open format .imzML files for downstream analysis. Transformation of data into the mass domain also provides greater accessibility, with mass information readily interpretable by users of established protein biology tools such as sodium dodecyl sulfate polyacrylamide gel electrophoresis.
中文翻译:

用于质量域分析的天然蛋白质质谱成像数据集的高通量解卷积
采用基于电喷雾的环境电离技术(例如纳喷雾解吸电喷雾电离 (nano-DESI))的蛋白质质谱成像 (MSI) 生成数据集,其中每个像素对应于由对应于多电荷蛋白质离子的峰填充的质谱。重要的是,与每种蛋白质相关的信号在多个电荷状态之间分配。这些峰可以通过光谱反卷积转换到质量域。当蛋白质在天然/非变性条件下成像以保留非共价相互作用时,反卷积对于帮助解释数据特别有价值。为了提高采集速度、信噪比和灵敏度,通常使用不提供同位素分辨率的质量分辨能力来执行原生 MSI,并且用于低分辨率数据去卷积的传统算法不适合这些大型数据集。UniDec 最初开发的目的是实现复杂蛋白质质谱的快速解卷积。在这里,我们开发了一个更新的功能集,利用高吞吐量模块 MetaUniDec 对本机 MSI 数据集的每个像素进行反卷积,并将m / z域图像文件转换为质量域。新工具可以读取、处理和输出开放格式的 .imzML 文件以进行下游分析。将数据转换为质量域还提供了更大的可访问性,使用已建立的蛋白质生物学工具(例如十二烷基硫酸钠聚丙烯酰胺凝胶电泳)的用户可以轻松解释质量信息。
更新日期:2023-09-06
中文翻译:

用于质量域分析的天然蛋白质质谱成像数据集的高通量解卷积
采用基于电喷雾的环境电离技术(例如纳喷雾解吸电喷雾电离 (nano-DESI))的蛋白质质谱成像 (MSI) 生成数据集,其中每个像素对应于由对应于多电荷蛋白质离子的峰填充的质谱。重要的是,与每种蛋白质相关的信号在多个电荷状态之间分配。这些峰可以通过光谱反卷积转换到质量域。当蛋白质在天然/非变性条件下成像以保留非共价相互作用时,反卷积对于帮助解释数据特别有价值。为了提高采集速度、信噪比和灵敏度,通常使用不提供同位素分辨率的质量分辨能力来执行原生 MSI,并且用于低分辨率数据去卷积的传统算法不适合这些大型数据集。UniDec 最初开发的目的是实现复杂蛋白质质谱的快速解卷积。在这里,我们开发了一个更新的功能集,利用高吞吐量模块 MetaUniDec 对本机 MSI 数据集的每个像素进行反卷积,并将m / z域图像文件转换为质量域。新工具可以读取、处理和输出开放格式的 .imzML 文件以进行下游分析。将数据转换为质量域还提供了更大的可访问性,使用已建立的蛋白质生物学工具(例如十二烷基硫酸钠聚丙烯酰胺凝胶电泳)的用户可以轻松解释质量信息。






























 京公网安备 11010802027423号
京公网安备 11010802027423号