当前位置:
X-MOL 学术
›
Anal. Methods
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Enhancing metabolite coverage in MALDI-MSI using laser post-ionisation (MALDI-2)
Analytical Methods ( IF 2.7 ) Pub Date : 2023-08-14 , DOI: 10.1039/d3ay01046e J C McKinnon 1 , H H Milioli 2, 3, 4 , C A Purcell 2, 3, 4 , C L Chaffer 2, 3, 4 , B Wadie 5 , T Alexandrov 5 , T W Mitchell 6 , S R Ellis 1
Analytical Methods ( IF 2.7 ) Pub Date : 2023-08-14 , DOI: 10.1039/d3ay01046e J C McKinnon 1 , H H Milioli 2, 3, 4 , C A Purcell 2, 3, 4 , C L Chaffer 2, 3, 4 , B Wadie 5 , T Alexandrov 5 , T W Mitchell 6 , S R Ellis 1
Affiliation
|
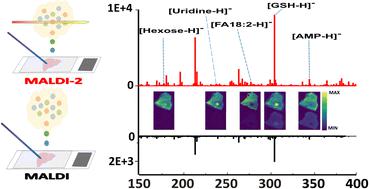
Matrix-assisted laser desorption/ionisation mass spectrometry imaging (MALDI-MSI) of metabolites can reveal how metabolism is altered throughout heterogeneous tissues. Here negative ion mode MALDI-MSI has been coupled with laser post-ionisation (MALDI-2) and applied to the MSI of low molecular weight (LMW) metabolites (<m/z 600) to investigate the benefits MALDI-2 offers for spatial metabolomics in terms of metabolite coverage and sensitivity. When applied to mouse kidney tissue MALDI-2 provided almost double the number of on-tissue specific mass features compared to conventional MALDI. MALDI-2 also resulted in not only the increased detection sensitivity for multiple metabolite species but also permitted the imaging of LMW metabolites (e.g. uridine) that were not detected using conventional MALDI-MSI. When compared against ∼140 publically available kidney datasets submitted through the METASPACE analysis platform using the same N-(1-naphthyl) ethylenediamine dihydrochloride (NEDC) matrix, MALDI-2 provided 34 unique metabolite m/z features that were not consistently annotated previously. To further evaluate the usefulness of this MALDI-2 approach to metabolite imaging, MALDI-2 was applied to the imaging of mouse liver tissue containing a metastasised breast cancer at a pixel size of 20 μm. Using a co-localisation analysis, MALDI-2 detected six tumour-specific metabolites that were not detected using conventional MALDI, as well as providing an up to 20-fold increase in signal intensities for many others (e.g., glutamate). This work provides one of the first reports of MALDI-2 applied to metabolite imaging and demonstrates the dramatic improvements in sensitivity and metabolite coverage it provides.
中文翻译:

使用激光后电离 (MALDI-2) 增强 MALDI-MSI 中的代谢物覆盖率
代谢物的基质辅助激光解吸/电离质谱成像 (MALDI-MSI) 可以揭示异质组织中代谢的变化情况。这里,负离子模式 MALDI-MSI 与激光后电离 (MALDI-2) 相结合,并应用于低分子量 (LMW) 代谢物 (< m / z 600 ) 的 MSI,以研究 MALDI-2 在空间方面的优势代谢组学的代谢物覆盖率和敏感性。当应用于小鼠肾组织时,MALDI-2 提供的组织特异性质量特征数量几乎是传统 MALDI 的两倍。MALDI-2 不仅提高了多种代谢物种类的检测灵敏度,而且还允许对传统 MALDI-MSI 无法检测到的LMW 代谢物(例如尿苷)进行成像。当与使用相同的N- (1-萘基)乙二胺二盐酸盐 (NEDC) 矩阵通过 METASPACE 分析平台提交的 ~140 个公开可用的肾脏数据集进行比较时,MALDI-2 提供了 34 个独特的代谢物m / z特征,这些特征之前未一致注释。为了进一步评估这种 MALDI-2 方法对代谢物成像的有用性,我们将 MALDI-2 用于对含有转移性乳腺癌的小鼠肝组织进行成像,像素大小为 20 μm。使用共定位分析,MALDI-2 检测到了使用传统 MALDI 无法检测到的六种肿瘤特异性代谢物,并且使许多其他代谢物(例如谷氨酸)的信号强度增加了高达 20倍。这项工作提供了 MALDI-2 应用于代谢物成像的首批报告之一,并展示了其在灵敏度和代谢物覆盖方面的显着改进。
更新日期:2023-08-14
中文翻译:

使用激光后电离 (MALDI-2) 增强 MALDI-MSI 中的代谢物覆盖率
代谢物的基质辅助激光解吸/电离质谱成像 (MALDI-MSI) 可以揭示异质组织中代谢的变化情况。这里,负离子模式 MALDI-MSI 与激光后电离 (MALDI-2) 相结合,并应用于低分子量 (LMW) 代谢物 (< m / z 600 ) 的 MSI,以研究 MALDI-2 在空间方面的优势代谢组学的代谢物覆盖率和敏感性。当应用于小鼠肾组织时,MALDI-2 提供的组织特异性质量特征数量几乎是传统 MALDI 的两倍。MALDI-2 不仅提高了多种代谢物种类的检测灵敏度,而且还允许对传统 MALDI-MSI 无法检测到的LMW 代谢物(例如尿苷)进行成像。当与使用相同的N- (1-萘基)乙二胺二盐酸盐 (NEDC) 矩阵通过 METASPACE 分析平台提交的 ~140 个公开可用的肾脏数据集进行比较时,MALDI-2 提供了 34 个独特的代谢物m / z特征,这些特征之前未一致注释。为了进一步评估这种 MALDI-2 方法对代谢物成像的有用性,我们将 MALDI-2 用于对含有转移性乳腺癌的小鼠肝组织进行成像,像素大小为 20 μm。使用共定位分析,MALDI-2 检测到了使用传统 MALDI 无法检测到的六种肿瘤特异性代谢物,并且使许多其他代谢物(例如谷氨酸)的信号强度增加了高达 20倍。这项工作提供了 MALDI-2 应用于代谢物成像的首批报告之一,并展示了其在灵敏度和代谢物覆盖方面的显着改进。















































 京公网安备 11010802027423号
京公网安备 11010802027423号