Med ( IF 12.8 ) Pub Date : 2023-07-07 , DOI: 10.1016/j.medj.2023.06.002 MacLean P Nasrallah 1 , Junhan Zhao 2 , Cheng Che Tsai 2 , David Meredith 3 , Eliana Marostica 4 , Keith L Ligon 5 , Jeffrey A Golden 6 , Kun-Hsing Yu 7
|
Background
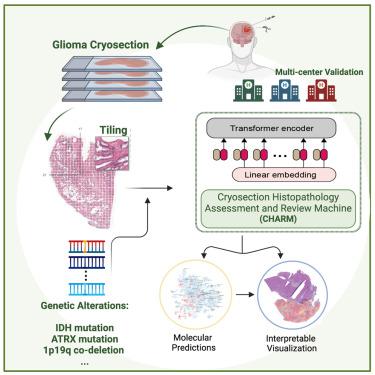
Timely and accurate intraoperative cryosection evaluations remain the gold standard for guiding surgical treatments for gliomas. However, the tissue-freezing process often generates artifacts that make histologic interpretation difficult. In addition, the 2021 WHO Classification of Tumors of the Central Nervous System incorporates molecular profiles in the diagnostic categories, so standard visual evaluation of cryosections alone cannot completely inform diagnoses based on the new classification system.
Methods
To address these challenges, we develop the context-aware Cryosection Histopathology Assessment and Review Machine (CHARM) using samples from 1,524 glioma patients from three different patient populations to systematically analyze cryosection slides.
Findings
Our CHARM models successfully identified malignant cells (AUROC = 0.98 ± 0.01 in the independent validation cohort), distinguished isocitrate dehydrogenase (IDH)-mutant tumors from wild type (AUROC = 0.79–0.82), classified three major types of molecularly defined gliomas (AUROC = 0.88–0.93), and identified the most prevalent subtypes of IDH-mutant tumors (AUROC = 0.89–0.97). CHARM further predicts clinically important genetic alterations in low-grade glioma, including ATRX, TP53, and CIC mutations, CDKN2A/B homozygous deletion, and 1p/19q codeletion via cryosection images.
Conclusions
Our approaches accommodate the evolving diagnostic criteria informed by molecular studies, provide real-time clinical decision support, and will democratize accurate cryosection diagnoses.
Funding
Supported in part by the National Institute of General Medical Sciences grant R35GM142879, the Google Research Scholar Award, the Blavatnik Center for Computational Biomedicine Award, the Partners’ Innovation Discovery Grant, and the Schlager Family Award for Early Stage Digital Health Innovations.
中文翻译:

冷冻切片病理学机器学习预测 2021 年 WHO 神经胶质瘤分类
背景
及时、准确的术中冷冻切片评估仍然是指导神经胶质瘤手术治疗的金标准。然而,组织冷冻过程经常产生伪影,使组织学解释变得困难。此外,2021 年世界卫生组织中枢神经系统肿瘤分类将分子谱纳入诊断类别,因此仅对冰冻切片进行标准视觉评估并不能完全为基于新分类系统的诊断提供信息。
方法
为了应对这些挑战,我们使用来自三个不同患者群体的 1,524 名神经胶质瘤患者的样本开发了情境感知冷冻切片组织病理学评估和审查机 (CHARM),以系统地分析冷冻切片。
发现
我们的 CHARM 模型成功识别了恶性细胞(独立验证队列中的 AUROC = 0.98 ± 0.01),将异柠檬酸脱氢酶 (IDH) 突变型肿瘤与野生型肿瘤区分开来 (AUROC = 0.79–0.82),将分子定义的神经胶质瘤分为三种主要类型 (AUROC = 0.88–0.93),并确定了 IDH 突变肿瘤最常见的亚型(AUROC = 0.89–0.97)。 CHARM 通过冷冻切片图像进一步预测低级别胶质瘤的临床重要遗传改变,包括ATRX 、 TP53和CIC突变、 CDKN2A/B纯合性缺失和 1p/19q 联合缺失。
结论
我们的方法适应分子研究不断发展的诊断标准,提供实时临床决策支持,并将使准确的冷冻切片诊断大众化。
资金
部分由美国国家普通医学科学研究所 R35GM142879 拨款、谷歌研究学者奖、布拉瓦尼克计算生物医学中心奖、合作伙伴创新发现拨款和施拉格家族早期数字健康创新奖提供支持。

































 京公网安备 11010802027423号
京公网安备 11010802027423号