背景
薰衣草链霉菌是一种产生 1-脱氧野尻霉素 (DNJ) 的细菌,DNJ 是一种具有多种有益药理特性的生物碱。虽然已经为这种细菌提出了推定的 DNJ 生物合成代谢途径,但相关的关键基因尚未确定。
结果
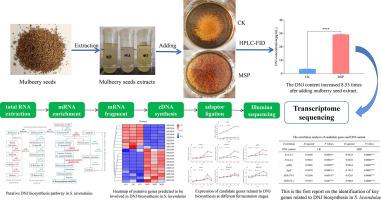
在这项研究中,桑籽提取物诱导了薰衣草发酵过程中的 DNJ 产生,并使用转录组测序鉴定了在这些实验条件下与 DNJ 生物合成相关的关键基因。然后使用实时 qPCR 分析基因表达,并与 DNJ 途径中的代谢物水平相关。桑籽多糖提取物 (MSP) 诱导薰衣草中的 DNJ 产量肉汤比桑籽生物碱和类黄酮提取物更有效。补充 3.00 mg/mL MSP 产生 27.19 μg/mL DNJ,是对照组(3.18 μg/mL)的 8.55 倍。转录组测序揭示了 MSP 与未补充对照比较中的 2328 个差异表达基因 (DEG),其中 1151 个 DEG 在 MSP 中上调,1177 个 DEG 下调。功能富集分析揭示了 DNJ 生物合成的四种途径。然后使用实时 qPCR 和中间代谢物分析验证与 DNJ 生物合成相关的六个关键基因。这些是,aspC,编码一种假定的转氨酶,产生 2-amino-2-deoxy- d -mannitol-6-phosphate;MJ0374-1和MJ0374-2, 编码产生 2-amino-2-deoxy- d -mannitol 的磷酸酶;和frmA-1、frmA-2和adhD,它们编码一种产生甘露尻霉素的锌依赖性脱氢酶。
结论
这是首次在薰衣草中鉴定出与 DNJ 生物合成相关的基因的研究,为通过工程改造细菌实现 DNJ 的工业规模生产铺平了道路。如何引用: Xin X, Jiang X, Niu B, et al. 淡紫色链霉菌1-脱氧野尻霉素生物合成相关关键基因的鉴定与表达。电子 J 生物技术 2023;64. https://doi.org/10.1016/j.ejbt.2023.03.003。
 "点击查看英文标题和摘要"
"点击查看英文标题和摘要"
Identification and expression of key genes related to 1-deoxynojirimycin biosynthesis in Streptomyces lavendulae
Background
Streptomyces lavendulae is a bacterium that produces 1-deoxynojirimycin (DNJ), an alkaloid with various beneficial pharmacological properties. While a putative DNJ biosynthesis metabolite pathway has been proposed for this bacterium, the related key genes have not yet been identified.
Results
In this study, DNJ production during S. lavendulae fermentation was induced by mulberry seed extracts and the key genes related to DNJ biosynthesis under those experimental conditions were identified using transcriptome sequencing. Gene expression was then analyzed using real-time qPCR, and related to metabolite levels in the DNJ pathway. The mulberry seed polysaccharide extract (MSP) induced DNJ yield in S. lavendulae broth more effectively than the mulberry seed alkaloid and flavonoid extracts. Supplementation with 3.00 mg/mL MSP resulted in 27.19 μg/mL DNJ, which was 8.55 times than that in the control group (3.18 μg/mL). Transcriptome sequencing revealed 2328 differentially expressed genes (DEGs) in MSP vs. unsupplemented-control comparisons, of which 1151 DEGs were up-regulated and 1177 DEGs were down-regulated in MSP. Functional enrichment analysis revealed four pathways of DNJ biosynthesis. Six key genes related to DNJ biosynthesis were then validated using real-time qPCR and the analysis of intermediate metabolites. These were, aspC, encoding a putative transaminase that produces 2-amino-2-deoxy-d-mannitol-6-phosphate; MJ0374-1 and MJ0374-2, encoding a phosphatase that produces 2-amino-2-deoxy-d-mannitol; and frmA-1, frmA-2, and adhD, which encode a Zn-dependent dehydrogenase that produces mannojirimycin.
Conclusions
This is the first study to identify the genes relevant for DNJ biosynthesis in S. lavendulae, and it paves the way for engineering the bacterium to achieve for industrial-scale DNJ production. How to cite: Xin X, Jiang X, Niu B, et al. Identification and expression of key genes related to 1-deoxynojirimycin biosynthesis in Streptomyces lavendulae. Electron J Biotechnol 2023; 64. https://doi.org/10.1016/j.ejbt.2023.03.003.
































 京公网安备 11010802027423号
京公网安备 11010802027423号