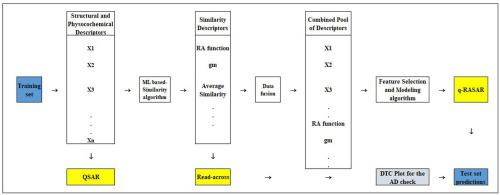
最近,通过使用各种机器学习(ML)引入了定量跨结构活性关系(q-RASAR)的概念 - 在传统的定量结构 - 活性关系(QSAR)建模框架中导出相似性函数,目的是在使用相同的可用化学信息内容的同时增强模型的外部预测性。本研究使用 hERG K +通道抑制心脏毒性,一个药学相关的终点,作为使用新的 q-RASAR 方法进行预测的模型集,因为该方法结合了 QSAR 和 Read-Across 的优点,并使用各种相似性和基于错误的方法生成简单且可解释的模型措施作为描述符。心脏毒性数据(根据 pIC50 值)是从文献中收集的。然后使用基于排序响应的划分算法将精选数据集分为训练集和测试集。重要的特征集是根据初始遗传算法模型的内部验证指标确定的。基于在最终多元线性回归 (MLR) 模型中选择的特征,RASAR 描述符是使用可从https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home 。然后将 RASAR 描述符与先前选择的特征合并,并使用网格搜索方法生成 MLR q-RASAR 模型。然后使用新颖的 DTC 适用性域图识别预测异常值,并在去除预测异常值后使用 q-RASAR 模型进行预测。生成最终的偏最小二乘(PLS) q-RASAR 模型以消除描述符之间的相互关联。还采用了其他各种机器学习方法,并基于交叉验证方法优化了相关超参数,并比较了最终的测试集预测结果。基于测试集预测和可解释性的表现,PLS q-RASAR 模型被选为最终模型,与先前报告的模型相比,即使不使用 3-D 描述符也能提供增强的预测能力。因此,该模型可用于分子的快速筛选,甚至在它们合成之前,以估计它们的心脏毒性潜力,从而确定分子的优先级,以便在药物发现管道中进行进一步的实验测试。还开发了一种基于 Java 的预测工具,用于快速筛选查询化合物的心脏毒性特性,并可从https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home。
 "点击查看英文标题和摘要"
"点击查看英文标题和摘要"
Machine-learning-based similarity meets traditional QSAR: “q-RASAR” for the enhancement of the external predictivity and detection of prediction confidence outliers in an hERG toxicity dataset
Recently, the concept of quantitative Read-Across Structure-Activity Relationship (q-RASAR) has been introduced by using various Machine Learning (ML) - derived similarity functions in the traditional quantitative structure-activity relationship (QSAR) modeling framework with the objective of enhancing the external predictivity of models while using the same available chemical information content. The present study uses the hERG K+ channel inhibition cardiotoxicity, a pharmaceutically relevant endpoint, as the modeling set for making predictions using the novel q-RASAR approach, as the approach combines the merits of QSAR and Read-Across, and generates simple and interpretable models using various similarity and error-based measures as descriptors. The cardiotoxicity data (in terms of pIC50 values) were collected from the literature. The curated data set was then divided into training and test sets using the sorted response-based division algorithm. The important set of features was identified based on the internal validation metrics of initial genetic algorithm models. Based on the features selected in the final Multiple Linear Regression (MLR) model, RASAR descriptors were computed using a tool available from https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home. The RASAR descriptors were then merged with the previously selected features, and an MLR q-RASAR model was generated using the grid search approach. The prediction outliers were then identified using the novel DTC Applicability Domain Plot, and the q-RASAR models were used for predictions after the removal of prediction outliers. A final Partial Least Squares (PLS) q-RASAR model was generated to obviate inter-correlation among descriptors. Various other Machine Learning approaches were also employed with the optimization of relevant hyperparameters based on the cross-validation approach, and the final test set prediction results were compared. Based on the performance in the test set predictions and interpretability, the PLS q-RASAR model was chosen as the final model which provided enhanced predictivity in comparison to previously reported models even without using 3-D descriptors. This model can thus be used for the quick screening of molecules, even before their synthesis, to estimate their cardiotoxic potential thus prioritizing molecules for further experimental testing in the drug discovery pipeline. A Java-based prediction tool has also been developed for the quick screening of cardiotoxic properties of query compounds and made available from https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home.


































 京公网安备 11010802027423号
京公网安备 11010802027423号