Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2022-12-26 , DOI: 10.1016/j.aca.2022.340748 Xia Cheng 1 , Xinyi Xia 2 , Dandan Ren 2 , Qiutong Chen 2 , Guanhong Xu 3 , Fangdi Wei 3 , Jing Yang 3 , Lin Wang 4 , Qin Hu 3 , Jianjun Zou 5 , Yao Cen 3
|
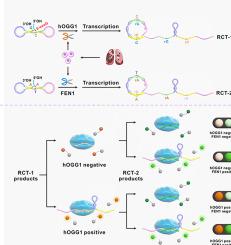
Human 8-oxoguanine DNA glycosylase (hOGG1) and flap endonuclease 1 (FEN1) are recognized as potential biomarkers in lung cancer investigations. Developing analytical platforms of simultaneously targeting hOGG1 and FEN1 with high selectivity, sensitivity, especially programmability and universality is highly valuable for clinical research. Herein, we established a signal-amplified platform for simultaneously detecting hOGG1 and FEN1 on the basis of cleavage-induced ligation of DNA dumbbell probes, rolling circle transcription (RCT) and CRISPR-Cas12a. A hOGG1 cleavable site and FEN1 cleavable flap were dexterously designed at the 5’ end of DNA flapped dumbbell probes (FDP) for hOGG1 and FEN1. After cleavage, the resulting nick sites with juxtaposition of 5′ phosphate and 3′ hydroxyl terminus could be linked to closed DNA dumbbell probes (CDP) by DNA ligase. The CDP served as a template for RCT, producing plentiful crRNA repeats to activate the trans-cleavage activity of CRISPR-Cas12a which could cleave fluorophores (TAMRA and FAM) and quenchers (BHQ2 and BHQ1) double-labeled ssDNA reporters. Then, hOGG1 and FEN1 could be detected by the recovered fluorescence signal, allowing for the highly sensitive calculated detection limits of 0.0013 and 0.0052 U/mL, respectively. Additionally, this method made it possible to evaluate the inhibitory effects, even to measure hOGG1 and FEN1 activities at the single-cell level. This novel target enzyme-initiated, circles-transcription without promoters, real-time generation, and self-assembly features of FDP-RCT-Cas12a system suppressed nonspecific background remarkably and relieved rigorous requirement of protospacer adjacent motif site. Hence, the universality of FDP-RCT-Cas12a system toward various disease-related non-nucleic acid targets which are tested without using aptamers was extremely improved.
中文翻译:

可编程 CRISPR-Cas12a 和自我招募 crRNA 辅助双生物传感平台,用于同时检测肺癌生物标志物 hOGG1 和 FEN1
人 8-氧代鸟嘌呤 DNA 糖基化酶 (hOGG1) 和瓣核酸内切酶 1 (FEN1) 被认为是肺癌研究中的潜在生物标志物。开发同时靶向 hOGG1 和 FEN1 的具有高选择性、灵敏度,尤其是可编程性和通用性的分析平台对临床研究具有很高的价值。在此,我们基于 DNA 哑铃探针的切割诱导连接、滚环转录 (RCT) 和 CRISPR-Cas12a,建立了同时检测 hOGG1 和FEN1的信号放大平台。hOGG1 可切割位点和 FEN1 可切割瓣被巧妙地设计在 DNA 的 5 '端flapped dumbbelp phOGG1 和 FEN1 的长袍 (FDP)。切割后,产生的具有 5' 磷酸和 3' 羟基末端并列的切口位点可以通过 DNA 连接酶连接到c losed DNA dumbbell probes (CDP)。CDP 作为 RCT 的模板,产生大量的 crRNA 重复以激活反式-CRISPR-Cas12a 的切割活性,可以切割荧光团(TAMRA 和 FAM)和淬灭剂(BHQ2 和 BHQ1)双标记 ssDNA 报告基因。然后,可以通过恢复的荧光信号检测到 hOGG1 和 FEN1,从而实现高度灵敏的计算检测限分别为 0.0013 和 0.0052 U/mL。此外,该方法还可以评估抑制作用,甚至可以在单细胞水平上测量 hOGG1 和 FEN1 的活性。FDP-RCT-Cas12a 系统的这种新型靶向酶启动、无启动子的环状转录、实时生成和自组装特征显着抑制了非特异性背景,并减轻了对原型间隔区相邻基序位点的严格要求。因此,

































 京公网安备 11010802027423号
京公网安备 11010802027423号