当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Bioinformatic Atlas of Radical SAM Enzyme-Modified RiPP Natural Products Reveals an Isoleucine–Tryptophan Crosslink
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2022-09-21 , DOI: 10.1021/jacs.2c06497 Kenzie A. Clark , Mohammad R. Seyedsayamdost
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2022-09-21 , DOI: 10.1021/jacs.2c06497 Kenzie A. Clark , Mohammad R. Seyedsayamdost
|
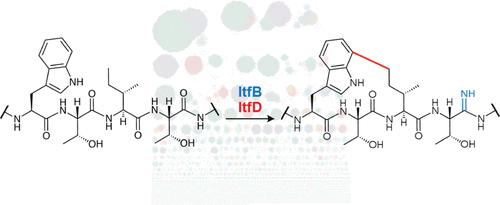
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a growing family of natural products with diverse activities and structures. RiPP classes are defined by the tailoring enzyme, which can introduce a narrow range of modifications or a diverse set of alterations. In the latter category, RiPPs synthesized by radical S-adenosylmethionine (SAM) enzymes, known as RaS-RiPPs, have emerged as especially divergent. A map of all RaS-RiPP gene clusters does not yet exist. Moreover, precursor peptides remain difficult to predict using computational methods. Herein, we have addressed these challenges and report a bioinformatic atlas of RaS-RiPP gene clusters in available microbial genome sequences. Using co-occurrence of RaS enzymes and transporters from varied families as a bioinformatic hook in conjunction with an in-house code to identify precursor peptides, we generated a map of ∼15,500 RaS-RiPP gene clusters, which reveal a remarkable diversity of syntenies pointing to a tremendous range of enzymatic and natural product chemistries that remain to be explored. To assess its utility, we examined one family of gene clusters encoding a YcaO enzyme and a RaS enzyme. We find the former is noncanonical, contains an iron–sulfur cluster, and installs a novel modification, a backbone amidine into the precursor peptide. The RaS enzyme was also found to install a new modification, a C–C crosslink between the unactivated terminal δ-methyl group of Ile and a Trp side chain. The co-occurrence search can be applied to other families of RiPPs, as we demonstrate with the emerging DUF692 di-iron enzyme superfamily.
中文翻译:

自由基SAM酶修饰的RiPP天然产物的生物信息学图谱揭示了异亮氨酸-色氨酸交联
核糖体合成和翻译后修饰肽 (RiPPs) 是一个不断增长的天然产物家族,具有不同的活性和结构。RiPP 类别由定制酶定义,它可以引入窄范围的修饰或多种改变。在后一类中,由自由基S合成的 RiPPs-腺苷甲硫氨酸 (SAM) 酶,称为 RaS-RiPPs,已经出现了特别不同的情况。尚不存在所有 RaS-RiPP 基因簇的图谱。此外,前体肽仍然难以使用计算方法进行预测。在这里,我们已经解决了这些挑战,并报告了可用微生物基因组序列中 RaS-RiPP 基因簇的生物信息图谱。使用来自不同家族的 RaS 酶和转运蛋白的共现作为生物信息学钩子,并结合内部代码来识别前体肽,我们生成了约 15,500 个 RaS-RiPP 基因簇的图谱,揭示了显着的同线性指向多样性有大量的酶和天然产物化学有待探索。为了评估其效用,我们检查了一个编码 YcaO 酶和 RaS 酶的基因簇家族。我们发现前者是非规范的,包含一个铁硫簇,并且在前体肽中安装了一种新的修饰,即骨架脒。还发现 RaS 酶安装了新的修饰,即 Ile 的未活化末端 δ-甲基和 Trp 侧链之间的 C-C 交联。正如我们用新兴的 DUF692 二铁酶超家族证明的那样,共现搜索可以应用于其他 RiPP 家族。
更新日期:2022-09-21
中文翻译:

自由基SAM酶修饰的RiPP天然产物的生物信息学图谱揭示了异亮氨酸-色氨酸交联
核糖体合成和翻译后修饰肽 (RiPPs) 是一个不断增长的天然产物家族,具有不同的活性和结构。RiPP 类别由定制酶定义,它可以引入窄范围的修饰或多种改变。在后一类中,由自由基S合成的 RiPPs-腺苷甲硫氨酸 (SAM) 酶,称为 RaS-RiPPs,已经出现了特别不同的情况。尚不存在所有 RaS-RiPP 基因簇的图谱。此外,前体肽仍然难以使用计算方法进行预测。在这里,我们已经解决了这些挑战,并报告了可用微生物基因组序列中 RaS-RiPP 基因簇的生物信息图谱。使用来自不同家族的 RaS 酶和转运蛋白的共现作为生物信息学钩子,并结合内部代码来识别前体肽,我们生成了约 15,500 个 RaS-RiPP 基因簇的图谱,揭示了显着的同线性指向多样性有大量的酶和天然产物化学有待探索。为了评估其效用,我们检查了一个编码 YcaO 酶和 RaS 酶的基因簇家族。我们发现前者是非规范的,包含一个铁硫簇,并且在前体肽中安装了一种新的修饰,即骨架脒。还发现 RaS 酶安装了新的修饰,即 Ile 的未活化末端 δ-甲基和 Trp 侧链之间的 C-C 交联。正如我们用新兴的 DUF692 二铁酶超家族证明的那样,共现搜索可以应用于其他 RiPP 家族。

































 京公网安备 11010802027423号
京公网安备 11010802027423号