Journal of Ethnopharmacology ( IF 4.8 ) Pub Date : 2022-08-17 , DOI: 10.1016/j.jep.2022.115630 Hai-Yan Jiang 1 , Hui-Yu Gao 2 , Jie Li 1 , Tian-Yu Zhou 3 , Shu-Ting Wang 1 , Jian-Bo Yang 2 , Rui-Rui Hao 1 , Fei Pang 1 , Feng Wei 2 , Zhi-Gang Liu 4 , Lian Kuang 1 , Shuang-Cheng Ma 2 , Jiu-Ming He 5 , Hong-Tao Jin 6
|
Ethnopharmacological relevance
The liver toxicity of Reynoutria multiflora (Thunb.) Moldenke. (Polygonaceae) (Polygonum multiflorum Thunb, PM) has always attracted much attention, but the related toxicity materials and mechanisms have not been elucidated due to multi-component and multi-target characteristics. In previous hepatotoxicity screening, different components of PM were first evaluated and the hepatotoxicity of component D [95% ethanol (EtOH) elution] in a 70% EtOH extract of PM (PM-D) showed the highest hepatotoxicity. Furthermore, the main components of PM-D were identified and their hepatotoxicity was evaluated based on a zebrafish embryo model. However, the hepatotoxicity mechanism of PM-D is unknown.
Aim of the study
This work is to explore the hepatotoxicity mechanisms of PM-D by integrating network toxicology and spatially resolved metabolomics strategy.
Materials and methods
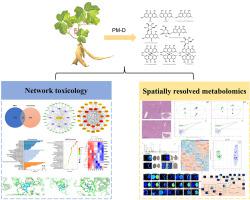
A hepatotoxicity interaction network of PM-D was constructed based on toxicity target prediction for eight key toxic ingredients and a hepatotoxicity target collection. Then the key signaling pathways were enriched, and molecular docking verification was implemented to evaluate the ability of toxic ingredients to bind to the core targets. The pathological changes of liver tissues and serum biochemical assays of mice were used to evaluate the liver injury effect of mice with oral administration of PM-D. Furthermore, spatially resolved metabolomics was used to visualize significant differences in metabolic profiles in mice after drug administration, to screen hepatotoxicity-related biomarkers and analyze metabolic pathways.
Results
The contents of four key toxic compounds in PM-D were detected. Network toxicology identified 30 potential targets of liver toxicity of PM-D. GO and KEGG enrichment analyses indicated that the hepatotoxicity of PM-D involved multiple biological activities, including cellular response to endogenous stimulus, organonitrogen compound metabolic process, regulation of the apoptotic process, regulation of kinase, regulation of reactive oxygen species metabolic process and signaling pathways including PI3K-Akt, AMPK, MAPK, mTOR, Ras and HIF-1. The molecular docking confirmed the high binding activity of 8 key toxic ingredients with 10 core targets, including mTOR, PIK3CA, AKT1, and EGFR. The high distribution of metabolites of PM-D in the liver of administrated mice was recognized by mass spectrometry imaging. Spatially resolved metabolomics results revealed significant changes in metabolic profiles after PM-D administration, and metabolites such as taurine, taurocholic acid, adenosine, and acyl-carnitines were associated with PM-D-induced liver injury. Enrichment analyses of metabolic pathways revealed tht linolenic acid and linoleic acid metabolism, carnitine synthesis, oxidation of branched-chain fatty acids, and six other metabolic pathways were significantly changed. Comprehensive analysis revealed that the hepatotoxicity caused by PM-D was closely related to cholestasis, mitochondrial damage, oxidative stress and energy metabolism, and lipid metabolism disorders.
Conclusions
In this study, the hepatotoxicity mechanisms of PM-D were comprehensively identified through an integrated spatially resolved metabolomics and network toxicology strategy, providing a theoretical foundation for the toxicity mechanisms of PM and its safe clinical application.
中文翻译:

综合空间分辨代谢组学和网络毒理学研究何首乌成分 D 的肝毒性机制
民族药理学相关性
Reynoutria multiflora (Thunb.) Moldenke的肝毒性。(Polygonaceae) ( Polygonum multiflorum Thunb, PM)一直备受关注,但由于其多成分、多靶点的特性,相关毒性材料及机制尚未阐明。在之前的肝毒性筛选中,首先评估了 PM 的不同成分,并且在 PM (PM-D) 的 70% EtOH 提取物中的成分 D [95% 乙醇 (EtOH) 洗脱] 的肝毒性显示出最高的肝毒性。此外,确定了 PM-D 的主要成分,并基于斑马鱼胚胎模型评估了它们的肝毒性。然而,PM-D 的肝毒性机制尚不清楚。
研究目的
这项工作是通过整合网络毒理学和空间分辨代谢组学策略来探索 PM-D 的肝毒性机制。
材料和方法
基于八种关键毒性成分的毒性目标预测和肝毒性目标集合,构建了PM-D的肝毒性相互作用网络。然后丰富关键信号通路,并进行分子对接验证,评估有毒成分与核心靶点的结合能力。采用小鼠肝组织病理变化和血清生化检测方法评价口服PM-D对小鼠的肝损伤作用。此外,空间分辨代谢组学用于观察给药后小鼠代谢特征的显着差异,筛选肝毒性相关生物标志物并分析代谢途径。
结果
检测了PM-D中四种关键有毒化合物的含量。网络毒理学确定了 PM-D 的 30 个潜在肝毒性目标。GO和KEGG富集分析表明,PM-D的肝毒性涉及多种生物活性,包括细胞对内源性刺激的反应、有机氮化合物代谢过程、凋亡过程的调节、激酶的调节、活性氧代谢过程的调节和信号通路包括PI3K-Akt、AMPK、MAPK、mTOR、Ras和HIF-1。分子对接证实了 8 种关键毒性成分与 10 个核心靶点的高结合活性,包括 mTOR、PIK3CA、AKT1 和 EGFR。质谱成像识别出给药小鼠肝脏中 PM-D 代谢物的高度分布。空间分辨代谢组学结果显示,PM-D 给药后代谢谱发生显着变化,牛磺酸、牛磺胆酸、腺苷和酰基肉碱等代谢物与 PM-D 诱导的肝损伤有关。代谢途径的富集分析揭示了亚麻酸和亚油酸代谢、肉碱合成、支链脂肪酸氧化等六种代谢途径发生显着变化。综合分析发现,PM-D引起的肝毒性与胆汁淤积、线粒体损伤、氧化应激和能量代谢、脂质代谢紊乱等密切相关。
结论
本研究通过整合空间分辨代谢组学和网络毒理学策略,全面鉴定PM-D的肝毒性机制,为PM的毒性机制及其临床安全应用提供理论基础。































 京公网安备 11010802027423号
京公网安备 11010802027423号