Pesticide Biochemistry and Physiology ( IF 4.2 ) Pub Date : 2022-07-28 , DOI: 10.1016/j.pestbp.2022.105181 Kangxu Wang 1 , Meiling Che 1 , Erhu Chen 1 , Fuji Jian 2 , Peian Tang 1
|
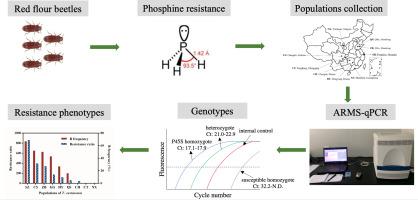
Resistance of Tribolium castaneum to phosphine is related to point mutations in DNA code corresponding to amino acid changes associated with a core metabolic enzyme dihydrolipoamide dehydrogenase (DLD), but the mutation patterns vary among different resistant populations. Thus, there is a great need to develop a cost-effective method to detect core mutations in T. castaneum, which would be the key factor to understand the molecular basis of phosphine resistance. Amplification refractory mutation system-based quantitative Real-Time PCR (ARMS-qPCR) is an ideal method that can rapidly detect point mutations. Here, the P45S and G131D mutations existed in the DLD of T. castaneum selected from strong Chinese resistance phenotypes, and the DLD P45S mutation, which represents a strong phosphine resistance allele, was confirmed as the most abundant mutation to determine strong resistance genotypes. Our study found that 85 out of 120 beetles carried the P45S resistance allele, including 51 homozygous and 34 heterozygous individuals. Moreover, there was a strong linear relationship (R2 = 0.917) between the resistance ratio and the resistance allele frequency among the strongly resistant populations. Our data showed that the ARMS-qPCR method that we developed could rapidly determine strong resistance phenotypes of T. castaneum to phosphine by detecting the DLD P45S mutation. These results not only provide a detailed example for developing an ARMS-qPCR-based method to characterize pesticide resistance, but also support further elucidation of the molecular basis of phosphine resistance.






























 京公网安备 11010802027423号
京公网安备 11010802027423号