Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2022-07-18 , DOI: 10.1016/j.csbj.2022.07.013 Samuel G Holmes 1, 2 , Balaji Nagarajan 1, 2 , Umesh R Desai 1, 2
|
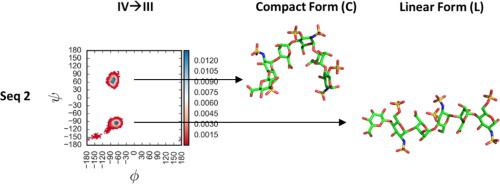
Heparan sulfate (HS) is arguably the most diverse linear biopolymer that is known to modulate hundreds of proteins. Whereas the configurational and conformational diversity of HS is well established in terms of varying sulfation patterns and iduronic acid (IdoA) puckers, a linear helical topology resembling a cylindrical rod is the only topology thought to be occupied by the biopolymer. We reasoned that 3-O-sulfation, a rare modification in natural HS, may induce novel topologies that contribute to selective recognition of proteins. In this work, we studied a library of 24 distinct HS hexasaccharides using molecular dynamics (MD). We discovered novel compact (C) topologies that are populated significantly by a unique group of 3-O-sulfated sequences containing IdoA residues. 3-O-sulfated sequences containing glucuronic acid (GlcA) residue and sequences devoid of 3-O-sulfate groups did not exhibit high levels of the C topology and primarily exhibited only the canonical linear (L) form. The C topology arises under dynamical conditions due to rotation around an IdoA→GlcN glycosidic linkage, especially in psi (Ψ) torsion. At an atomistic level, the L→C transformation is a multi-factorial phenomenon engineered to reduce like-charge repulsion, release one or more HS-bound water molecules, and organize a bi-dentate “IdoA-cation-IdoA” interaction. These forces also drive an L→C transformation in a 3-O-sulfated octasaccharide, which has shown evidence of the unique C topology in the co-crystallized state. The 3-O-sulfate-based generation of unique, sequence-specific, compact topologies indicate that natural HS encodes a dynamic sulfation code that could be exploited for selective recognition of target proteins.
中文翻译:

3-O-硫酸化在硫酸乙酰肝素中诱导序列特异性紧凑型拓扑,编码动态硫酸化代码
硫酸乙酰肝素 (HS) 可以说是最多样化的线性生物聚合物,已知可调节数百种蛋白质。尽管 HS 的构型和构象多样性在不同的硫酸化模式和艾杜糖醛酸 (IdoA) 褶皱方面得到了很好的证实,但类似于圆柱棒的线性螺旋拓扑结构是唯一被认为被生物聚合物占据的拓扑结构。我们推断 3- O-硫酸化是天然 H2S 中的一种罕见修饰,可能会诱导有助于选择性识别蛋白质的新型拓扑结构。在这项工作中,我们使用分子动力学 (MD) 研究了一个包含 24 种不同 HS 六糖的库。我们发现了新颖的紧凑 (C) 拓扑结构,这些拓扑结构由一组独特的包含 IdoA 残基的 3- O-硫酸化序列显着填充。3-含有葡萄糖醛酸 (GlcA) 残基的O-硫酸化序列和不含 3- O-硫酸基团的序列没有表现出高水平的 C 拓扑结构,主要仅表现出典型的线性 (L) 形式。由于围绕 IdoA→GlcN 糖苷键旋转,C 拓扑在动力学条件下出现,尤其是在 psi (Ψ) 扭转中。在原子水平上,L→C 转变是一种多因素现象,旨在减少同种电荷的排斥,释放一个或多个与 HS 结合的水分子,并组织双齿“IdoA-阳离子-IdoA”相互作用。这些力还驱动 3- O-硫酸化八糖中的 L→C 转变,这已显示共结晶状态下独特 C 拓扑结构的证据。3- O- 基于硫酸盐生成的独特的、序列特异性的、紧凑的拓扑结构表明天然 H2S 编码动态硫酸化代码,可用于选择性识别目标蛋白质。































 京公网安备 11010802027423号
京公网安备 11010802027423号