Food Research International ( IF 7.0 ) Pub Date : 2022-05-10 , DOI: 10.1016/j.foodres.2022.111344 Yucong Zou 1 , Xiaofeng Li 1 , Xuan Xin 2 , Haixia Xu 1 , Lan Mo 1 , Yigang Yu 1 , Guanglei Zhao 3
|
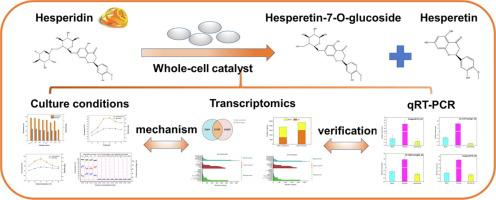
一种新的黑曲霉培养全细胞催化剂用于橙皮苷(HES)的级联水解,以生产高转化率(90%以上)的高价值橙皮素-7-O-葡萄糖苷(HG)和橙皮素。此外,所使用的诱导剂被证明可用于细胞生长并诱导细胞产生特定的酶。值得注意的是,诱导剂的类型决定了细胞是否可以水解 HES。产品组成也可以通过调整不同的诱导剂来控制。转录组分析表明柚皮苷-vs-空白组和蔗糖-vs-空白组的基因表达存在明显差异。柚皮苷-vs-空白组主要是上调的差异表达基因(DEGs),而蔗糖-vs-空白组主要是下调的差异表达基因(DEGs)。基因本体论(GO)分析表明,无论是加入柚皮苷还是蔗糖作为诱导剂,都会极大地影响细胞的催化活性。此外,发现了3个与鼠李糖苷酶相关的基因、14个与葡萄糖苷酶相关的基因和5个与水解酶活性相关的基因。这些基因不仅参与鼠李糖苷酶和葡萄糖苷酶的活性,而且还参与剪接体以及蔗糖和淀粉代谢途径。定量实时聚合酶链反应(qRT-PCR)分析表明转录组测序结果可靠。该研究提供了一种水解 HES 的新方法,以及理解与全细胞催化剂水解相关的机制的新视角。发现了14个与葡萄糖苷酶相关的基因和5个与水解酶活性相关的基因。这些基因不仅参与鼠李糖苷酶和葡萄糖苷酶的活性,而且还参与剪接体以及蔗糖和淀粉代谢途径。定量实时聚合酶链反应(qRT-PCR)分析表明转录组测序结果可靠。该研究提供了一种水解 HES 的新方法,以及理解与全细胞催化剂水解相关的机制的新视角。发现了14个与葡萄糖苷酶相关的基因和5个与水解酶活性相关的基因。这些基因不仅参与鼠李糖苷酶和葡萄糖苷酶的活性,而且还参与剪接体以及蔗糖和淀粉代谢途径。定量实时聚合酶链反应(qRT-PCR)分析表明转录组测序结果可靠。该研究提供了一种水解 HES 的新方法,以及理解与全细胞催化剂水解相关的机制的新视角。定量实时聚合酶链反应(qRT-PCR)分析表明转录组测序结果可靠。该研究提供了一种水解 HES 的新方法,以及理解与全细胞催化剂水解相关的机制的新视角。定量实时聚合酶链反应(qRT-PCR)分析表明转录组测序结果可靠。该研究提供了一种水解 HES 的新方法,以及理解与全细胞催化剂水解相关的机制的新视角。

"点击查看英文标题和摘要"


















































 京公网安备 11010802027423号
京公网安备 11010802027423号